A Triple Gene-Deleted Pseudorabies Virus-Vectored Subunit PCV2b and CSFV Vaccine Protects Pigs against PCV2b Challenge and Induces Serum Neutralizing Antibody Response against CSFV
- PMID: 35214763
- PMCID: PMC8878206
- DOI: 10.3390/vaccines10020305
A Triple Gene-Deleted Pseudorabies Virus-Vectored Subunit PCV2b and CSFV Vaccine Protects Pigs against PCV2b Challenge and Induces Serum Neutralizing Antibody Response against CSFV
Abstract
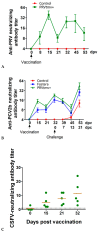
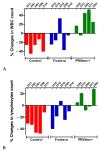
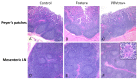
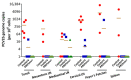
Porcine circovirus type 2 (PCV2) is endemic worldwide. PCV2 causes immunosuppressive infection. Co-infection of pigs with other swine viruses, such as pseudorabies virus (PRV) and classical swine fever virus (CSFV), have fatal outcomes, causing the swine industry significant economic losses in many if not all pig-producing countries. Currently available inactivated/modified-live/vectored vaccines against PCV2/CSFV/PRV have safety and efficacy limitations. To address these shortcomings, we have constructed a triple gene (thymidine kinase, glycoprotein E [gE], and gG)-deleted (PRVtmv) vaccine vector expressing chimeric PCV2b-capsid, CSFV-E2, and chimeric Erns-fused with bovine granulocytic monocyte-colony stimulating factor (Erns-GM-CSF), designated as PRVtmv+, a trivalent vaccine. Here we compared this vaccine's immunogenicity and protective efficacy in pigs against wild-type PCV2b challenge with that of the inactivated Zoetis Fostera Gold PCV commercial vaccine. The live PRVtmv+ prototype trivalent subunit vaccine is safe and highly attenuated in pigs. Based on PCV2b-specific neutralizing antibody titers, viremia, viral load in lymphoid tissues, fecal-virus shedding, and leukocyte/lymphocyte count, the PRVtmv+ yielded better protection for vaccinated pigs than the commercial vaccine after the PCV2b challenge. Additionally, the PRVtmv+ vaccinated pigs generated low to moderate levels of CSFV-specific neutralizing antibodies.
Keywords: CSFV; PCV2 capsid; PRV trivalent vaccine; glycoproteins E2 and Erns; granulocytic monocyte-colony stimulating factor (GM-CSF); pig; pseudorabies virus; triple mutant; vaccine efficacy; vectored vaccine.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
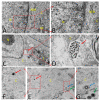
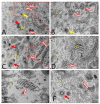




Similar articles
-
Live Triple Gene-Deleted Pseudorabies Virus-Vectored Subunit PCV2b and CSFV Vaccine Undergoes an Abortive Replication Cycle in the TG Neurons following Latency Reactivation.Viruses. 2023 Feb 8;15(2):473. doi: 10.3390/v15020473. Viruses. 2023. PMID: 36851689 Free PMC article.
-
A Triple Gene-Deleted Pseudorabies Virus-Vectored Subunit PCV2b and CSFV Vaccine Protect Pigs against a Virulent CSFV Challenge.Viruses. 2023 Oct 25;15(11):2143. doi: 10.3390/v15112143. Viruses. 2023. PMID: 38005821 Free PMC article.
-
Generation and Immunogenicity of a Recombinant Pseudorabies Virus Co-Expressing Classical Swine Fever Virus E2 Protein and Porcine Circovirus Type 2 Capsid Protein Based on Fosmid Library Platform.Pathogens. 2019 Dec 1;8(4):279. doi: 10.3390/pathogens8040279. Pathogens. 2019. PMID: 31805703 Free PMC article.
-
Use of interleukin 12 to enhance the cellular immune response of swine to an inactivated herpesvirus vaccine.Adv Vet Med. 1999;41:447-61. doi: 10.1016/s0065-3519(99)80034-2. Adv Vet Med. 1999. PMID: 9890035 Review.
-
Laboratory diagnosis, epizootiology, and efficacy of marker vaccines in classical swine fever: a review.Vet Q. 2000 Oct;22(4):182-8. doi: 10.1080/01652176.2000.9695054. Vet Q. 2000. PMID: 11087126 Review.
Cited by
-
Live Triple Gene-Deleted Pseudorabies Virus-Vectored Subunit PCV2b and CSFV Vaccine Undergoes an Abortive Replication Cycle in the TG Neurons following Latency Reactivation.Viruses. 2023 Feb 8;15(2):473. doi: 10.3390/v15020473. Viruses. 2023. PMID: 36851689 Free PMC article.
-
A Novel Quadruple Gene-Deleted BoHV-1-Vectored RVFV Subunit Vaccine Induces Humoral and Cell-Mediated Immune Response against Rift Valley Fever in Calves.Viruses. 2023 Oct 30;15(11):2183. doi: 10.3390/v15112183. Viruses. 2023. PMID: 38005861 Free PMC article.
-
A Triple Gene-Deleted Pseudorabies Virus-Vectored Subunit PCV2b and CSFV Vaccine Protect Pigs against a Virulent CSFV Challenge.Viruses. 2023 Oct 25;15(11):2143. doi: 10.3390/v15112143. Viruses. 2023. PMID: 38005821 Free PMC article.
References
-
- Ellis J.A., Bratanich A., Clark E.G., Allan G., Meehan B., Haines D.M., Harding J., West K.H., Krakowka S., Konoby C., et al. Coinfection by porcine circoviruses and porcine parvovirus in pigs with naturally acquired postweaning multisystemic wasting syndrome. J. Vet. Diagn. Investig. 2000;12:21–27. doi: 10.1177/104063870001200104. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

