Characterization of the TCR β Chain Repertoire in Peripheral Blood from Hepatitis B Vaccine Responders and Non-Responders
- PMID: 35210805
- PMCID: PMC8856041
- DOI: 10.2147/JIR.S347702
Characterization of the TCR β Chain Repertoire in Peripheral Blood from Hepatitis B Vaccine Responders and Non-Responders
Abstract
Background: Hepatitis B (HepB) vaccination can effectively prevent the prevalence of hepatitis B virus (HBV) infection. However, the incidence of vaccination failure is about 5~10% and the underlying molecular mechanisms are poorly understood. T cells have an essential role in the recipient's immune response to vaccine, which could be elucidated by high-throughput sequencing (HTS) and bioinformatics analysis.
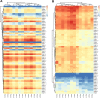
Methods: We conducted HTS of the T cell receptor β chain (TRB) complementarity-determining region 3 (CDR3) repertoires in eighteen positive responders (responders) and 10 negative responders (non-responders) who all had HepB vaccination, the repertoire features of BV, BJ and V-J genes and their diversity, respectively, were compared between the positive and negative responders using the Mann-Whitney test. Moreover, the relatively conserved motifs in CDR3 were revealed and compared to those in the other group's report.
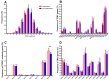
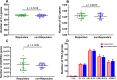
Results: The diversity of TRB CDR3 and the frequencies of BV27 and BV7-9 are significantly increased for HepB vaccine responders compared to those in non-responders. The motifs of CDR3s in BV27/J1-1, BV27/J2-5, and BV7-9/J2-5, respectively, were most expressed as "NTE", "QETQ", and "GG-Q (E)-ETQ". Moreover, the motif "KLNSPL" was determined in nearly 80% CDR3s in BV27/J1-6 from HepB vaccine responders for the first time.
Conclusion: Our results present the comprehensive profiles of TRB CDR3 in the HepB vaccine responders and non-responders after standard vaccination protocol and determine the relatively conservative motifs of CDR3s that may respond to the HepB vaccine. Further results suggest that the profile of TRB repertoire could distinguish the HepB vaccine responders from non-responders and provide a new target for optimizing and improving the efficiency of the HepB vaccine.
Keywords: complementarity-determining region 3; hepatitis B surface antibody; hepatitis B vaccination; high-throughput sequencing; immune repertoire.
© 2022 Yang et al.
Conflict of interest statement
The authors report no conflicts of interest for this work.
Figures


Similar articles
-
Next-generation sequencing analysis of the human T-cell and B-cell receptor repertoire diversity before and after hepatitis B vaccination.Hum Vaccin Immunother. 2019;15(11):2738-2753. doi: 10.1080/21645515.2019.1600987. Epub 2019 Jul 25. Hum Vaccin Immunother. 2019. PMID: 30945971 Free PMC article.
-
Profiling T cell receptor β-chain in responders after immunization with recombinant hepatitis B vaccine.J Gene Med. 2021 Sep;23(9):e3367. doi: 10.1002/jgm.3367. Epub 2021 Jun 17. J Gene Med. 2021. PMID: 34048625
-
Antibody response to revaccination among adult non-responders to primary Hepatitis B vaccination in China.Hum Vaccin Immunother. 2015;11(11):2716-22. doi: 10.1080/21645515.2015.1045172. Epub 2015 Aug 7. Hum Vaccin Immunother. 2015. PMID: 26252481 Free PMC article.
-
The status of hepatitis B control in the African region.Pan Afr Med J. 2017 Jun 22;27(Suppl 3):17. doi: 10.11604/pamj.supp.2017.27.3.11981. eCollection 2017. Pan Afr Med J. 2017. PMID: 29296152 Free PMC article. Review.
-
Ruminant livestock TR V(D)J genes and CDR3 repertoire.Vet Immunol Immunopathol. 2024 Nov;277:110829. doi: 10.1016/j.vetimm.2024.110829. Epub 2024 Sep 22. Vet Immunol Immunopathol. 2024. PMID: 39316948 Review.
Cited by
-
Upregulated CD8+ MAIT cell differentiation and KLRD1 gene expression after inactivated SARS-CoV-2 vaccination identified by single-cell sequencing.Front Immunol. 2023 Aug 15;14:1174406. doi: 10.3389/fimmu.2023.1174406. eCollection 2023. Front Immunol. 2023. PMID: 37654490 Free PMC article.
-
T-Cell Receptor β Chain and B-Cell Receptor Repertoires in Chronic Hepatitis B Patients with Coexisting HBsAg and Anti-HBs.Pathogens. 2022 Jun 26;11(7):727. doi: 10.3390/pathogens11070727. Pathogens. 2022. PMID: 35889974 Free PMC article.
-
T-Cell Receptor Repertoire Sequencing and Its Applications: Focus on Infectious Diseases and Cancer.Int J Mol Sci. 2022 Aug 2;23(15):8590. doi: 10.3390/ijms23158590. Int J Mol Sci. 2022. PMID: 35955721 Free PMC article. Review.
-
The T-cell receptor β chain CDR3 insights of bovine liver immune repertoire under heat stress.Anim Biosci. 2024 Dec;37(12):2178-2188. doi: 10.5713/ab.24.0152. Epub 2024 Jun 25. Anim Biosci. 2024. PMID: 38938039 Free PMC article.
-
Similar usage of T-cell receptor β-chain between tumor and adjacent normal tissue in hepatocellular carcinoma.Cancer Med. 2024 Aug;13(16):e70121. doi: 10.1002/cam4.70121. Cancer Med. 2024. PMID: 39192502 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

