Structural basis of adenylyl cyclase 9 activation
- PMID: 35210418
- PMCID: PMC8873477
- DOI: 10.1038/s41467-022-28685-y
Structural basis of adenylyl cyclase 9 activation
Abstract
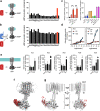
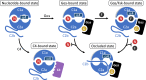
Adenylyl cyclase 9 (AC9) is a membrane-bound enzyme that converts ATP into cAMP. The enzyme is weakly activated by forskolin, fully activated by the G protein Gαs subunit and is autoinhibited by the AC9 C-terminus. Although our recent structural studies of the AC9-Gαs complex provided the framework for understanding AC9 autoinhibition, the conformational changes that AC9 undergoes in response to activator binding remains poorly understood. Here, we present the cryo-EM structures of AC9 in several distinct states: (i) AC9 bound to a nucleotide inhibitor MANT-GTP, (ii) bound to an artificial activator (DARPin C4) and MANT-GTP, (iii) bound to DARPin C4 and a nucleotide analogue ATPαS, (iv) bound to Gαs and MANT-GTP. The artificial activator DARPin C4 partially activates AC9 by binding at a site that overlaps with the Gαs binding site. Together with the previously observed occluded and forskolin-bound conformations, structural comparisons of AC9 in the four conformations described here show that secondary structure rearrangements in the region surrounding the forskolin binding site are essential for AC9 activation.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Insights into the Regulatory Properties of Human Adenylyl Cyclase Type 9.Mol Pharmacol. 2019 Apr;95(4):349-360. doi: 10.1124/mol.118.114595. Epub 2019 Jan 29. Mol Pharmacol. 2019. PMID: 30696718 Free PMC article.
-
Regulatory sites of CaM-sensitive adenylyl cyclase AC8 revealed by cryo-EM and structural proteomics.EMBO Rep. 2024 Mar;25(3):1513-1540. doi: 10.1038/s44319-024-00076-y. Epub 2024 Feb 13. EMBO Rep. 2024. PMID: 38351373 Free PMC article.
-
Structural basis for the inhibition of mammalian membrane adenylyl cyclase by 2 '(3')-O-(N-Methylanthraniloyl)-guanosine 5 '-triphosphate.J Biol Chem. 2005 Feb 25;280(8):7253-61. doi: 10.1074/jbc.M409076200. Epub 2004 Dec 9. J Biol Chem. 2005. PMID: 15591060
-
The chilling of adenylyl cyclase 9 and its translational potential.Cell Signal. 2020 Jun;70:109589. doi: 10.1016/j.cellsig.2020.109589. Epub 2020 Feb 24. Cell Signal. 2020. PMID: 32105777 Review.
-
Guanyl nucleotide regulation of hormonally-responsive adenylyl cyclases.Mol Cell Endocrinol. 1979 Dec;16(3):129-46. doi: 10.1016/0303-7207(79)90022-4. Mol Cell Endocrinol. 1979. PMID: 230102 Review.
Cited by
-
Biochemical pharmacology of adenylyl cyclases in cancer.Biochem Pharmacol. 2024 Oct;228:116160. doi: 10.1016/j.bcp.2024.116160. Epub 2024 Mar 24. Biochem Pharmacol. 2024. PMID: 38522554 Review.
-
Role and Interplay of Different Signaling Pathways Involved in Sciatic Nerve Regeneration.J Mol Neurosci. 2024 Nov 12;74(4):108. doi: 10.1007/s12031-024-02286-4. J Mol Neurosci. 2024. PMID: 39531101 Review.
-
Human adenylyl cyclase 9 is auto-stimulated by its isoform-specific C-terminal domain.Life Sci Alliance. 2023 Jan 19;6(4):e202201791. doi: 10.26508/lsa.202201791. Print 2023 Apr. Life Sci Alliance. 2023. PMID: 36657828 Free PMC article.
-
The evolutionary conservation of eukaryotic membrane-bound adenylyl cyclase isoforms.Front Pharmacol. 2022 Sep 27;13:1009797. doi: 10.3389/fphar.2022.1009797. eCollection 2022. Front Pharmacol. 2022. PMID: 36238545 Free PMC article.
-
Comparative Analyses Reveal the Genetic Mechanism of Ambergris Production in the Sperm Whale Based on the Chromosome-Level Genome.Animals (Basel). 2023 Jan 20;13(3):361. doi: 10.3390/ani13030361. Animals (Basel). 2023. PMID: 36766250 Free PMC article.
References
-
- Chao QI, S. S, Medalia O, Korkhov VM. The structure of a membrane adenylyl cyclase bound to an activated stimulatory G protein. Science. 2019;364:389–394. - PubMed
-
- Pierre S, Eschenhagen T, Geisslinger G, Scholich K. Capturing adenylyl cyclases as potential drug targets. Nat. Rev. Drug Discov. 2009;8:321–335. - PubMed
-
- Khannpnavar B, Mehta V, Qi C, Korkhov V. Structure and function of adenylyl cyclases, key enzymes in cellular signaling. Curr. Opin. Struct. Biol. 2020;63:34–41. - PubMed
-
- Tang WJ, Gilman AG. Adenylyl cyclases. Cell. 1992;70:869–872. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

