The Removal of Time-Concentration Data Points from Progress Curves Improves the Determination of Km: The Example of Paraoxonase 1
- PMID: 35209091
- PMCID: PMC8874660
- DOI: 10.3390/molecules27041306
The Removal of Time-Concentration Data Points from Progress Curves Improves the Determination of Km: The Example of Paraoxonase 1
Abstract
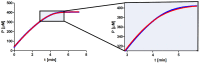
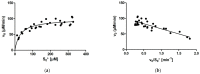
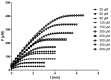
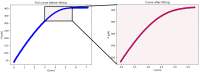
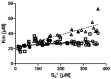
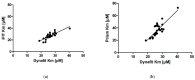
Several approaches for determining an enzyme's kinetic parameter Km (Michaelis constant) from progress curves have been developed in recent decades. In the present article, we compare different approaches on a set of experimental measurements of lactonase activity of paraoxonase 1 (PON1): (1) a differential-equation-based Michaelis-Menten (MM) reaction model in the program Dynafit; (2) an integrated MM rate equation, based on an approximation of the Lambert W function, in the program GraphPad Prism; (3) various techniques based on initial rates; and (4) the novel program "iFIT", based on a method that removes data points outside the area of maximum curvature from the progress curve, before analysis with the integrated MM rate equation. We concluded that the integrated MM rate equation alone does not determine kinetic parameters precisely enough; however, when coupled with a method that removes data points (e.g., iFIT), it is highly precise. The results of iFIT are comparable to the results of Dynafit and outperform those of the approach with initial rates or with fitting the entire progress curve in GraphPad Prism; however, iFIT is simpler to use and does not require inputting a reaction mechanism. Removing unnecessary points from progress curves and focusing on the area around the maximum curvature is highly advised for all researchers determining Km values from progress curves.
Keywords: Lambert W; dihydrocoumarin; integrated Michaelis–Menten equation; lactonase activity; paraoxonase 1; progress curves.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


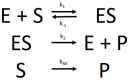
Similar articles
-
iFIT: An automated web tool for determining enzyme-kinetic parameters based on the high-curvature region of progress curves.Acta Chim Slov. 2022 Jun 14;69(2):478-482. doi: 10.17344/acsi.2022.7359. Acta Chim Slov. 2022. PMID: 35861063
-
Evaluation of the paraoxonase-1 kinetic parameters of the lactonase activity by nonlinear fit of progress curves.J Enzyme Inhib Med Chem. 2020 Dec;35(1):261-264. doi: 10.1080/14756366.2019.1695792. J Enzyme Inhib Med Chem. 2020. PMID: 31790606 Free PMC article.
-
The reliability of Michaelis constants and maximum velocities estimated by using the integrated Michaelis-Menten equation.Biochem J. 1973 Dec;135(4):779-84. doi: 10.1042/bj1350779. Biochem J. 1973. PMID: 4778274 Free PMC article.
-
Validity of the Michaelis-Menten equation--steady-state or reactant stationary assumption: that is the question.FEBS J. 2014 Jan;281(2):464-72. doi: 10.1111/febs.12564. Epub 2013 Nov 18. FEBS J. 2014. PMID: 24245583 Review.
-
High performance enzyme kinetics of turnover, activation and inhibition for translational drug discovery.Expert Opin Drug Discov. 2017 Jan;12(1):17-37. doi: 10.1080/17460441.2017.1245721. Epub 2016 Nov 3. Expert Opin Drug Discov. 2017. PMID: 27784173 Review.
Cited by
-
Vutiglabridin Modulates Paraoxonase 1 and Ameliorates Diet-Induced Obesity in Hyperlipidemic Mice.Biomolecules. 2023 Apr 18;13(4):687. doi: 10.3390/biom13040687. Biomolecules. 2023. PMID: 37189434 Free PMC article.
-
Investigation of Paraoxonase-1 Genotype and Enzyme-Kinetic Parameters in the Context of Cognitive Impairment in Parkinson's Disease.Antioxidants (Basel). 2023 Feb 7;12(2):399. doi: 10.3390/antiox12020399. Antioxidants (Basel). 2023. PMID: 36829958 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

