The Ribosomal Protein RpL22 Interacts In Vitro with 5'-UTR Sequences Found in Some Drosophila melanogaster Transposons
- PMID: 35205350
- PMCID: PMC8872304
- DOI: 10.3390/genes13020305
The Ribosomal Protein RpL22 Interacts In Vitro with 5'-UTR Sequences Found in Some Drosophila melanogaster Transposons
Abstract
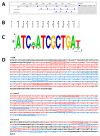
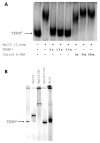
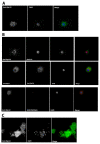
Mobility of eukaryotic transposable elements (TEs) are finely regulated to avoid an excessive mutational load caused by their movement. The transposition of retrotransposons is usually regulated through the interaction of host- and TE-encoded proteins, with non-coding regions (LTR and 5'-UTR) of the transposon. Examples of new potent cis-acting sequences, identified and characterized in the non-coding regions of retrotransposons, include the insulator of gypsy and Idefix, and the enhancer of ZAM of Drosophila melanogaster. Recently we have shown that in the 5'-UTR of the LTR-retrotransposon ZAM there is a sequence structured in tandem-repeat capable of operating as an insulator both in Drosophila (S2R+) and human cells (HEK293). Here, we test the hypothesis that tandem repeated 5'-UTR of a different LTR-retrotransposon could accommodate similar regulatory elements. The comparison of the 5'-UTR of some LTR-transposons allowed us to identify a shared motif of 13 bp, called Transposable Element Redundant Motif (TERM). Surprisingly, we demonstrated, by Yeast One-Hybrid assay, that TERM interacts with the D. melanogaster ribosomal protein RpL22. The Drosophila RpL22 has additional Ala-, Lys- and Pro-rich sequences at the amino terminus, which resembles the carboxy-terminal portion of histone H1 and histone H5. For this reason, it has been hypothesized that RpL22 might have two functions, namely the role in organizing the ribosome, and a potential regulatory role involving DNA-binding similar to histone H1, which represses transcription in Drosophila. In this paper, we show, by two independent sets of experiments, that DmRpL22 is able to directly and specifically bind DNA of Drosophila melanogaster.
Keywords: DNA-protein interaction; Drosophila; Rpl22; histone 1-like; ribosomal protein; transposable elements.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures

Similar articles
-
Evidences for insulator activity of the 5'UTR of the Drosophila melanogaster LTR-retrotransposon ZAM.Mol Genet Genomics. 2010 May;283(5):503-9. doi: 10.1007/s00438-010-0529-4. Epub 2010 Apr 3. Mol Genet Genomics. 2010. PMID: 20364351
-
Heterochromatin protein 1 interacts with 5'UTR of transposable element ZAM in a sequence-specific fashion.Gene. 2007 May 15;393(1-2):1-10. doi: 10.1016/j.gene.2006.12.028. Epub 2007 Jan 19. Gene. 2007. PMID: 17343996
-
Evidence of the Physical Interaction between Rpl22 and the Transposable Element Doc5, a Heterochromatic Transposon of Drosophila melanogaster.Genes (Basel). 2021 Dec 16;12(12):1997. doi: 10.3390/genes12121997. Genes (Basel). 2021. PMID: 34946947 Free PMC article.
-
Mechanisms of LTR-Retroelement Transposition: Lessons from Drosophila melanogaster.Viruses. 2017 Apr 16;9(4):81. doi: 10.3390/v9040081. Viruses. 2017. PMID: 28420154 Free PMC article. Review.
-
Impact of multiple insertions of two retroelements, ZAM and Idefix at an euchromatic locus.Genetica. 2000;109(1-2):53-9. doi: 10.1023/a:1026534207401. Genetica. 2000. PMID: 11293795 Review.
Cited by
-
The Stability and Evolution of Genes and Genomes.Genes (Basel). 2023 Aug 31;14(9):1747. doi: 10.3390/genes14091747. Genes (Basel). 2023. PMID: 37761887 Free PMC article.
-
The Green Valley of Drosophila melanogaster Constitutive Heterochromatin: Protein-Coding Genes Involved in Cell Division Control.Cells. 2022 Sep 29;11(19):3058. doi: 10.3390/cells11193058. Cells. 2022. PMID: 36231024 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

