Synthesis and Structure-Activity Relationships of Novel Non-Steroidal CYP17A1 Inhibitors as Potential Prostate Cancer Agents
- PMID: 35204665
- PMCID: PMC8961587
- DOI: 10.3390/biom12020165
Synthesis and Structure-Activity Relationships of Novel Non-Steroidal CYP17A1 Inhibitors as Potential Prostate Cancer Agents
Abstract
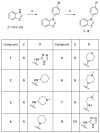
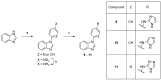
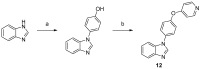
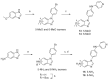
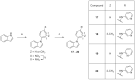
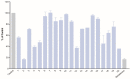
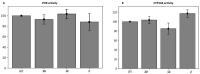
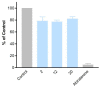
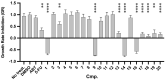
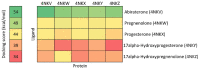
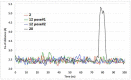
Twenty new compounds, targeting CYP17A1, were synthesized, based on our previous work on a benzimidazole scaffold, and their biological activity evaluated. Inhibition of CYP17A1 is an important modality in the treatment of prostate cancer, which remains the most abundant cancer type in men. The biological assessment included CYP17A1 hydroxylase and lyase inhibition, CYP3A4 and P450 oxidoreductase (POR) inhibition, as well as antiproliferative activity in PC3 prostate cancer cells. The most potent compounds were selected for further analyses including in silico modeling. This combined effort resulted in a compound (comp 2, IC50 1.2 µM, in CYP17A1) with a potency comparable to abiraterone and selectivity towards the other targets tested. In addition, the data provided an understanding of the structure-activity relationship of this novel non-steroidal compound class.
Keywords: CYP17A1; cytochrome P450 17A1; enzyme inhibition; prostate cancer.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures
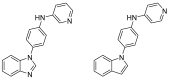
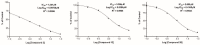
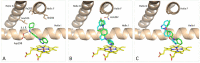
Similar articles
-
Discovery of Novel Non-Steroidal Cytochrome P450 17A1 Inhibitors as Potential Prostate Cancer Agents.Int J Mol Sci. 2020 Jul 9;21(14):4868. doi: 10.3390/ijms21144868. Int J Mol Sci. 2020. PMID: 32660148 Free PMC article.
-
Promising Tools in Prostate Cancer Research: Selective Non-Steroidal Cytochrome P450 17A1 Inhibitors.Sci Rep. 2016 Jul 12;6:29468. doi: 10.1038/srep29468. Sci Rep. 2016. PMID: 27406023 Free PMC article.
-
A-ring modified steroidal azoles retaining similar potent and slowly reversible CYP17A1 inhibition as abiraterone.J Steroid Biochem Mol Biol. 2014 Sep;143:1-10. doi: 10.1016/j.jsbmb.2014.01.013. Epub 2014 Feb 6. J Steroid Biochem Mol Biol. 2014. PMID: 24508512 Free PMC article.
-
CYP17A1: a biochemistry, chemistry, and clinical review.Curr Top Med Chem. 2013;13(12):1364-84. doi: 10.2174/1568026611313120002. Curr Top Med Chem. 2013. PMID: 23688130 Review.
-
17alpha-hydroxylase/17,20-lyase (p450(17alpha)) inhibitors in the treatment of prostate cancer: a review.Anticancer Agents Med Chem. 2009 Jul;9(6):613-26. doi: 10.2174/187152009788680046. Anticancer Agents Med Chem. 2009. PMID: 19601745 Review.
Cited by
-
Exploring the Chemical Space of CYP17A1 Inhibitors Using Cheminformatics and Machine Learning.Molecules. 2023 Feb 9;28(4):1679. doi: 10.3390/molecules28041679. Molecules. 2023. PMID: 36838665 Free PMC article.
-
Effect of Essential Oil Components on the Activity of Steroidogenic Cytochrome P450.Biomolecules. 2024 Feb 8;14(2):203. doi: 10.3390/biom14020203. Biomolecules. 2024. PMID: 38397440 Free PMC article.
-
Exploring the Potential of Sulfur Moieties in Compounds Inhibiting Steroidogenesis.Biomolecules. 2023 Sep 5;13(9):1349. doi: 10.3390/biom13091349. Biomolecules. 2023. PMID: 37759751 Free PMC article.
-
How Is CYP17A1 Activity Altered in Autism? A Pilot Study to Identify Potential Pharmacological Targets.Life (Basel). 2022 Jun 10;12(6):867. doi: 10.3390/life12060867. Life (Basel). 2022. PMID: 35743898 Free PMC article.
-
Non-steroidal CYP17A1 Inhibitors: Discovery and Assessment.J Med Chem. 2023 May 25;66(10):6542-6566. doi: 10.1021/acs.jmedchem.3c00442. Epub 2023 May 16. J Med Chem. 2023. PMID: 37191389 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

