Plasma-Derived HIV-1 Virions Contain Considerable Levels of Defective Genomes
- PMID: 35201897
- PMCID: PMC8941922
- DOI: 10.1128/jvi.02011-21
Plasma-Derived HIV-1 Virions Contain Considerable Levels of Defective Genomes
Abstract
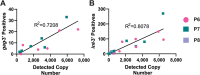
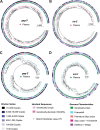
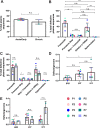
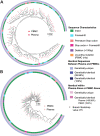
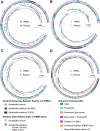
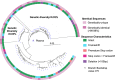
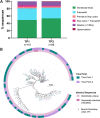
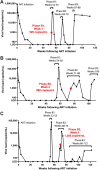
Genetically-characterizing full-length HIV-1 RNA is critical for identifying genetically-intact genomes and for comparing these RNA genomes to proviral DNA. We have developed a method for sequencing plasma-derived RNA using long-range sequencing (PRLS assay; ∼8.3 kb from gag to the 3' end or ∼5 kb from integrase to the 3' end). We employed the gag-3' PRLS assay to sequence HIV-1 RNA genomes from ART-naive participants during acute/early infection (n = 6) or chronic infection (n = 2). On average, only 65% of plasma-derived genomes were genetically-intact. Defects were found in all genomic regions but were concentrated in env and pol. We compared these genomes to near-full-length proviral sequences from paired peripheral blood mononuclear cell (PBMC) samples for the acute/early group and found that near-identical (>99.98% identical) sequences were identified only during acute infection. For three participants who initiated therapy during acute infection, we used the int-3' PRLS assay to sequence plasma-derived genomes from an analytical treatment interruption and identified 100% identical genomes between pretherapy and rebound time points. The PRLS assay provides a new level of sensitivity for understanding the genetic composition of plasma-derived HIV-1 RNA from viremic individuals either pretherapy or after treatment interruption, which will be invaluable in assessing possible HIV-1 curative strategies. IMPORTANCE We developed novel plasma-derived RNA using long-range sequencing assays (PRLS assay; 8.3 kb, gag-3', and 5.0 kb, int-3'). Employing the gag-3' PRLS assay, we found that 26% to 51% of plasma-derived genomes are genetically-defective, largely as a result of frameshift mutations and deletions. These genetic defects were concentrated in the env region compared to gag and pol, likely a reflection of viral immune escape in env during untreated HIV-1 infection. Employing the int-3' PRLS assay, we found that analytical treatment interruption (ATI) plasma-derived sequences were identical and genetically-intact. Several sequences from the ATI plasma samples were identical to viral sequences from pretherapy plasma and PBMC samples, indicating that HIV-1 reservoirs established prior to therapy contribute to viral rebound during an ATI. Therefore, near-full-length sequencing of HIV-1 particles is required to gain an accurate picture of the genetic landscape of plasma HIV-1 virions in studies of HIV-1 replication and persistence.
Keywords: human immunodeficiency virus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Intact HIV Proviruses Persist in Children Seven to Nine Years after Initiation of Antiretroviral Therapy in the First Year of Life.J Virol. 2020 Jan 31;94(4):e01519-19. doi: 10.1128/JVI.01519-19. Print 2020 Jan 31. J Virol. 2020. PMID: 31776265 Free PMC article.
-
Discordance between HIV-1 Population in Plasma at Rebound after Structured Treatment Interruption and Archived Provirus Population in Peripheral Blood Mononuclear Cells.Microbiol Spectr. 2022 Aug 31;10(4):e0135322. doi: 10.1128/spectrum.01353-22. Epub 2022 Jun 14. Microbiol Spectr. 2022. PMID: 35699458 Free PMC article.
-
Impact of Antiretroviral Therapy Duration on HIV-1 Infection of T Cells within Anatomic Sites.J Virol. 2020 Jan 17;94(3):e01270-19. doi: 10.1128/JVI.01270-19. Print 2020 Jan 17. J Virol. 2020. PMID: 31723024 Free PMC article.
-
Single-molecule techniques to quantify and genetically characterise persistent HIV.Retrovirology. 2018 Jan 9;15(1):3. doi: 10.1186/s12977-017-0386-x. Retrovirology. 2018. PMID: 29316955 Free PMC article. Review.
-
New Approaches to Multi-Parametric HIV-1 Genetics Using Multiple Displacement Amplification: Determining the What, How, and Where of the HIV-1 Reservoir.Viruses. 2021 Dec 10;13(12):2475. doi: 10.3390/v13122475. Viruses. 2021. PMID: 34960744 Free PMC article. Review.
Cited by
-
CD4+ T cells with latent HIV-1 have reduced proliferative responses to T cell receptor stimulation.J Exp Med. 2024 Mar 4;221(3):e20231511. doi: 10.1084/jem.20231511. Epub 2024 Jan 25. J Exp Med. 2024. PMID: 38270554 Free PMC article.
-
Full-spectrum HIV drug resistance mutation detection by high-resolution complete pol gene sequencing.J Clin Virol. 2023 Jul;164:105491. doi: 10.1016/j.jcv.2023.105491. Epub 2023 May 6. J Clin Virol. 2023. PMID: 37182384 Free PMC article.
-
Unequal distribution of genetically-intact HIV-1 proviruses in cells expressing the immune checkpoint markers PD-1 and/or CTLA-4.Front Immunol. 2023 Jan 26;14:1064346. doi: 10.3389/fimmu.2023.1064346. eCollection 2023. Front Immunol. 2023. PMID: 36776833 Free PMC article.
-
New Assay Reveals Vast Excess of Defective over Intact HIV-1 Transcripts in Antiretroviral Therapy-Suppressed Individuals.J Virol. 2022 Dec 21;96(24):e0160522. doi: 10.1128/jvi.01605-22. Epub 2022 Nov 30. J Virol. 2022. PMID: 36448806 Free PMC article.
-
Understanding latent HIV-1 reservoirs through host genomics approaches.iScience. 2023 Oct 28;26(11):108342. doi: 10.1016/j.isci.2023.108342. eCollection 2023 Nov 17. iScience. 2023. PMID: 38026212 Free PMC article. Review.
References
-
- Finzi D, Hermankova M, Pierson T, Carruth LM, Buck C, Chaisson RE, Quinn TC, Chadwick K, Margolick J, Brookmeyer R, Gallant J, Markowitz M, Ho DD, Richman DD, Siliciano RF. 1997. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278:1295–1300. 10.1126/science.278.5341.1295. - DOI - PubMed
-
- Chun TW, Justement JS, Murray D, Hallahan CW, Maenza J, Collier AC, Sheth PM, Kaul R, Ostrowski M, Moir S, Kovacs C, Fauci AS. 2010. Rebound of plasma viremia following cessation of antiretroviral therapy despite profoundly low levels of HIV reservoir: implications for eradication. AIDS 24:2803–2808. 10.1097/QAD.0b013e328340a239. - DOI - PMC - PubMed
-
- Davey RT, Bhat N, Yoder C, Chun T-W, Metcalf JA, Dewar R, Natarajan V, Lempicki RA, Adelsberger JW, Miller KD, Kovacs JA, Polis MA, Walker RE, Falloon J, Masur H, Gee D, Baseler M, Dimitrov DS, Fauci AS, Lane HC. 1999. HIV-1 and T cell dynamics after interruption of highly active antiretroviral therapy (HAART) in patients with a history of sustained viral suppression. Proc Natl Acad Sci U S A 96:15109–15114. 10.1073/pnas.96.26.15109. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

