Microfluidic Compartmentalization Platforms for Single Cell Analysis
- PMID: 35200319
- PMCID: PMC8869497
- DOI: 10.3390/bios12020058
Microfluidic Compartmentalization Platforms for Single Cell Analysis
Abstract
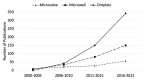
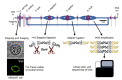
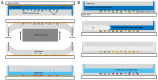
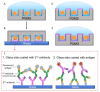
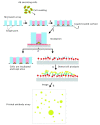
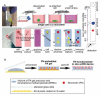
Many cellular analytical technologies measure only the average response from a cell population with an assumption that a clonal population is homogenous. The ensemble measurement often masks the difference among individual cells that can lead to misinterpretation. The advent of microfluidic technology has revolutionized single-cell analysis through precise manipulation of liquid and compartmentalizing single cells in small volumes (pico- to nano-liter). Due to its advantages from miniaturization, microfluidic systems offer an array of capabilities to study genomics, transcriptomics, and proteomics of a large number of individual cells. In this regard, microfluidic systems have emerged as a powerful technology to uncover cellular heterogeneity and expand the depth and breadth of single-cell analysis. This review will focus on recent developments of three microfluidic compartmentalization platforms (microvalve, microwell, and microdroplets) that target single-cell analysis spanning from proteomics to genomics. We also compare and contrast these three microfluidic platforms and discuss their respective advantages and disadvantages in single-cell analysis.
Keywords: droplets; microvalves; microwells; single-cell analysis; single-cell compartmentalization.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
A design and optimization of a high throughput valve based microfluidic device for single cell compartmentalization and analysis.Sci Rep. 2021 Jun 21;11(1):12995. doi: 10.1038/s41598-021-92472-w. Sci Rep. 2021. PMID: 34155296 Free PMC article.
-
Development of Droplet Microfluidics Enabling High-Throughput Single-Cell Analysis.Molecules. 2016 Jul 5;21(7):881. doi: 10.3390/molecules21070881. Molecules. 2016. PMID: 27399651 Free PMC article. Review.
-
A facile single-cell patterning strategy based on harbor-like microwell microfluidics.Biomed Mater. 2024 May 31;19(4). doi: 10.1088/1748-605X/ad4e83. Biomed Mater. 2024. PMID: 38772387
-
Advancing single-cell proteomics and metabolomics with microfluidic technologies.Analyst. 2019 Jan 28;144(3):846-858. doi: 10.1039/c8an01503a. Analyst. 2019. PMID: 30351310 Review.
-
Microfluidic techniques for high throughput single cell analysis.Curr Opin Biotechnol. 2016 Aug;40:90-96. doi: 10.1016/j.copbio.2016.02.015. Epub 2016 Mar 28. Curr Opin Biotechnol. 2016. PMID: 27032065 Free PMC article. Review.
Cited by
-
Inference of B cell clonal families using heavy/light chain pairing information.PLoS Comput Biol. 2022 Nov 28;18(11):e1010723. doi: 10.1371/journal.pcbi.1010723. eCollection 2022 Nov. PLoS Comput Biol. 2022. PMID: 36441808 Free PMC article.
-
Microfluidic design in single-cell sequencing and application to cancer precision medicine.Cell Rep Methods. 2023 Sep 25;3(9):100591. doi: 10.1016/j.crmeth.2023.100591. Epub 2023 Sep 18. Cell Rep Methods. 2023. PMID: 37725985 Free PMC article. Review.
-
A Microfluidic Approach for Probing Heterogeneity in Cytotoxic T-Cells by Cell Pairing in Hydrogel Droplets.Micromachines (Basel). 2022 Nov 4;13(11):1910. doi: 10.3390/mi13111910. Micromachines (Basel). 2022. PMID: 36363930 Free PMC article.
-
Research progress in isolation and identification of rumen probiotics.Front Cell Infect Microbiol. 2024 May 21;14:1411482. doi: 10.3389/fcimb.2024.1411482. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 38836057 Free PMC article. Review.
-
Recent Approaches to Design and Analysis of Electrical Impedance Systems for Single Cells Using Machine Learning.Sensors (Basel). 2023 Jun 28;23(13):5990. doi: 10.3390/s23135990. Sensors (Basel). 2023. PMID: 37447838 Free PMC article. Review.

