Generation of CRISPR-Cas9-mediated genetic knockout human intestinal tissue-derived enteroid lines by lentivirus transduction and single-cell cloning
- PMID: 35197604
- PMCID: PMC9059808
- DOI: 10.1038/s41596-021-00669-0
Generation of CRISPR-Cas9-mediated genetic knockout human intestinal tissue-derived enteroid lines by lentivirus transduction and single-cell cloning
Abstract
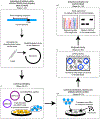
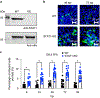
Human intestinal tissue-derived enteroids (HIEs; also called organoids) are a powerful ex vivo model for gastrointestinal research. Genetic modification of these nontransformed cultures allows new insights into gene function and biological processes involved in intestinal diseases as well as gastrointestinal and donor segment-specific function. Here we provide a detailed technical pipeline and protocol for using the CRISPR-Cas9 genome editing system to knock out a gene of interest specifically in HIEs by lentiviral transduction and single-cell cloning. This protocol differs from a previously published alternative using electroporation of human colonoids to deliver piggyback transposons or CRISPR-Cas9 constructs, as this protocol uses a modified, fused LentiCRISPRv2-small-guiding RNA to express Cas9 and small-guiding RNA in a lentivirus. The protocol also includes the steps of gene delivery and subsequent single-cell cloning of the knockout cells as well as verification of clones and sequence identification of the mutation sites to establish knockout clones. An overview flowchart, step-by-step guidelines and troubleshooting suggestions are provided to aid the researcher in obtaining the genetic knockout HIE line within 2-3 months. In this protocol, we further describe how to use HIEs as an ex vivo model to assess host restriction factors for viral replication (using human norovirus replication as an example) by knocking out host attachment factors or innate immunity genes. Other applications are discussed to broaden the utility of this system, for example, to generate knockin or conditional knockout HIE lines to investigate the function of essential genes in many biological processes including other types of organoids.
© 2022. The Author(s), under exclusive licence to Springer Nature Limited.
Figures



Similar articles
-
CRISPR/Cas9-Mediated Gene Knockout in Cells and Tissues Using Lentivirus.Curr Protoc. 2023 May;3(5):e772. doi: 10.1002/cpz1.772. Curr Protoc. 2023. PMID: 37222511 Free PMC article.
-
Development of Knock-Out Muscle Cell Lines using Lentivirus-Mediated CRISPR/Cas9 Gene Editing.J Vis Exp. 2022 Jun 16;(184). doi: 10.3791/64114. J Vis Exp. 2022. PMID: 35781470
-
A Robust Protocol for CRISPR-Cas9 Gene Editing in Human Suspension Cell Lines.Curr Protoc. 2021 Nov;1(11):e286. doi: 10.1002/cpz1.286. Curr Protoc. 2021. PMID: 34748280
-
Versatile and precise gene-targeting strategies for functional studies in mammalian cell lines.Methods. 2017 May 15;121-122:45-54. doi: 10.1016/j.ymeth.2017.05.003. Epub 2017 May 10. Methods. 2017. PMID: 28499832 Review.
-
Primary Airway Epithelial Cell Gene Editing Using CRISPR-Cas9.Methods Mol Biol. 2018;1706:267-292. doi: 10.1007/978-1-4939-7471-9_15. Methods Mol Biol. 2018. PMID: 29423804 Review.
Cited by
-
Gastrointestinal organoids in the study of viral infections.Am J Physiol Gastrointest Liver Physiol. 2023 Jan 1;324(1):G51-G59. doi: 10.1152/ajpgi.00152.2022. Epub 2022 Nov 22. Am J Physiol Gastrointest Liver Physiol. 2023. PMID: 36414538 Free PMC article. Review.
-
Recent advances in therapeutic CRISPR-Cas9 genome editing: mechanisms and applications.Mol Biomed. 2023 Apr 7;4(1):10. doi: 10.1186/s43556-023-00115-5. Mol Biomed. 2023. PMID: 37027099 Free PMC article. Review.
-
Applications of 3D organoids in toxicological studies: a comprehensive analysis based on bibliometrics and advances in toxicological mechanisms.Arch Toxicol. 2024 Aug;98(8):2309-2330. doi: 10.1007/s00204-024-03777-4. Epub 2024 May 28. Arch Toxicol. 2024. PMID: 38806717 Review.
-
Motion lubrication suppressed mechanical activation via hydrated fibrous gene patch for tendon healing.Sci Adv. 2023 Feb 10;9(6):eadc9375. doi: 10.1126/sciadv.adc9375. Epub 2023 Feb 10. Sci Adv. 2023. PMID: 36763658 Free PMC article.
-
Organoids in gastrointestinal diseases: from experimental models to clinical translation.Gut. 2022 Sep;71(9):1892-1908. doi: 10.1136/gutjnl-2021-326560. Epub 2022 May 30. Gut. 2022. PMID: 35636923 Free PMC article. Review.
References
-
- Sato T et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology 141, 1762–1772 (2011). - PubMed
-
- Middendorp S et al. Adult stem cells in the small intestine are intrinsically programmed with their location-specific function. Stem Cells 32, 1083–1091 (2014). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

