Spread of Gamma (P.1) Sub-Lineages Carrying Spike Mutations Close to the Furin Cleavage Site and Deletions in the N-Terminal Domain Drives Ongoing Transmission of SARS-CoV-2 in Amazonas, Brazil
- PMID: 35196783
- PMCID: PMC8865440
- DOI: 10.1128/spectrum.02366-21
Spread of Gamma (P.1) Sub-Lineages Carrying Spike Mutations Close to the Furin Cleavage Site and Deletions in the N-Terminal Domain Drives Ongoing Transmission of SARS-CoV-2 in Amazonas, Brazil
Abstract
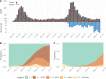
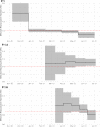
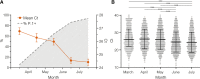
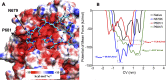
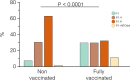
The Amazonas was one of the most heavily affected Brazilian states by the COVID-19 epidemic. Despite a large number of infected people, particularly during the second wave associated with the spread of the Variant of Concern (VOC) Gamma (lineage P.1), SARS-CoV-2 continues to circulate in the Amazonas. To understand how SARS-CoV-2 persisted in a human population with a high immunity barrier, we generated 1,188 SARS-CoV-2 whole-genome sequences from individuals diagnosed in the Amazonas state from 1st January to 6th July 2021, of which 38 were vaccine breakthrough infections. Our study reveals a sharp increase in the relative prevalence of Gamma plus (P.1+) variants, designated Pango Lineages P.1.3 to P.1.6, harboring two types of additional Spike changes: deletions in the N-terminal (NTD) domain (particularly Δ144 or Δ141-144) associated with resistance to anti-NTD neutralizing antibodies or mutations at the S1/S2 junction (N679K or P681H) that probably enhance the binding affinity to the furin cleavage site, as suggested by our molecular dynamics simulations. As lineages P.1.4 (S:N679K) and P.1.6 (S:P681H) expanded (Re > 1) from March to July 2021, the lineage P.1 declined (Re < 1) and the median Ct value of SARS-CoV-2 positive cases in Amazonas significantly decreases. Still, we did not find an increased incidence of P.1+ variants among breakthrough cases of fully vaccinated patients (71%) in comparison to unvaccinated individuals (93%). This evidence supports that the ongoing endemic transmission of SARS-CoV-2 in the Amazonas is driven by the spread of new local Gamma/P.1 sublineages that are more transmissible, although not more efficient to evade vaccine-elicited immunity than the parental VOC. Finally, as SARS-CoV-2 continues to spread in human populations with a declining density of susceptible hosts, the risk of selecting more infectious variants or antibody evasion mutations is expected to increase. IMPORTANCE The continuous evolution of SARS-CoV-2 is an expected phenomenon that will continue to happen due to the high number of cases worldwide. The present study analyzed how a Variant of Concern (VOC) could still circulate in a population hardly affected by two COVID-19 waves and with vaccination in progress. Our results showed that the answer behind that was a new generation of Gamma-like viruses, which emerged locally carrying mutations that made it more transmissible and more capable of spreading, partially evading prior immunity triggered by natural infections or vaccines. With thousands of new cases daily, the current pandemics scenario suggests that SARS-CoV-2 will continue to evolve and efforts to reduce the number of infected subjects, including global equitable access to COVID-19 vaccines, are mandatory. Thus, until the end of pandemics, the SARS-CoV-2 genomic surveillance will be an essential tool to better understand the drivers of the viral evolutionary process.
Keywords: Brazil; COVID-19; SARS-CoV-2; coronavirus; variant gamma; virus evolution.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Re-emergence of Gamma-like-II and emergence of Gamma-S:E661D SARS-CoV-2 lineages in the south of Brazil after the 2021 outbreak.Virol J. 2021 Nov 17;18(1):222. doi: 10.1186/s12985-021-01690-1. Virol J. 2021. PMID: 34789293 Free PMC article.
-
Genomic Surveillance of SARS-CoV-2 Lineages Indicates Early Circulation of P.1 (Gamma) Variant of Concern in Southern Brazil.Microbiol Spectr. 2022 Feb 23;10(1):e0151121. doi: 10.1128/spectrum.01511-21. Epub 2022 Feb 16. Microbiol Spectr. 2022. PMID: 35171035 Free PMC article.
-
The P681H Mutation in the Spike Glycoprotein of the Alpha Variant of SARS-CoV-2 Escapes IFITM Restriction and Is Necessary for Type I Interferon Resistance.J Virol. 2022 Dec 14;96(23):e0125022. doi: 10.1128/jvi.01250-22. Epub 2022 Nov 9. J Virol. 2022. PMID: 36350154 Free PMC article.
-
The Biological Functions and Clinical Significance of SARS-CoV-2 Variants of Corcern.Front Med (Lausanne). 2022 May 20;9:849217. doi: 10.3389/fmed.2022.849217. eCollection 2022. Front Med (Lausanne). 2022. PMID: 35669924 Free PMC article. Review.
-
Roles of the polybasic furin cleavage site of spike protein in SARS-CoV-2 replication, pathogenesis, and host immune responses and vaccination.J Med Virol. 2022 May;94(5):1815-1820. doi: 10.1002/jmv.27539. Epub 2021 Dec 31. J Med Virol. 2022. PMID: 34936124 Review.
Cited by
-
New variants of COVID-19 (XBB.1.5 and XBB.1.16, the "Arcturus"): A review of highly questioned concerns, a brief comparison between different peaks in the COVID-19 pandemic, with a focused systematic review on expert recommendations for prevention, vaccination, and treatment measures in the general population and at-risk groups.Immun Inflamm Dis. 2024 Jun;12(6):e1323. doi: 10.1002/iid3.1323. Immun Inflamm Dis. 2024. PMID: 38938013 Free PMC article. Review.
-
Immunogenicity, Effectiveness, and Safety of Inactivated Virus (CoronaVac) Vaccine in a Two-Dose Primary Protocol and BNT162b2 Heterologous Booster in Brazil (Immunita-001): A One Year Period Follow Up Phase 4 Study.Front Immunol. 2022 Jun 9;13:918896. doi: 10.3389/fimmu.2022.918896. eCollection 2022. Front Immunol. 2022. PMID: 35757764 Free PMC article. Clinical Trial.
-
Haplotypic Distribution of SARS-CoV-2 Variants in Cases of Intradomiciliary Infection in the State of Rondônia, Western Amazon.Bioinform Biol Insights. 2024 Nov 21;18:11779322241266354. doi: 10.1177/11779322241266354. eCollection 2024. Bioinform Biol Insights. 2024. PMID: 39574519 Free PMC article.
-
Implementing a provisional overarching intervention for COVID-19 monitoring and control in the Brazil-Colombia-Peru frontier.Front Public Health. 2024 Jan 8;11:1330347. doi: 10.3389/fpubh.2023.1330347. eCollection 2023. Front Public Health. 2024. PMID: 38259793 Free PMC article.
-
Epidemiological and Clinical Features of SARS-CoV-2 Variants Circulating between April-December 2021 in Italy.Viruses. 2022 Nov 12;14(11):2508. doi: 10.3390/v14112508. Viruses. 2022. PMID: 36423117 Free PMC article.
References
-
- Fundação de Vigilância em Saúde do Amazonas. 2021. Boletim diaria COVID-19 no Amazonas 15–06-2021. https://www.fvs.am.gov.br/noticias_view/6264. Accessed July 03, 2021.
-
- Naveca FG, Nascimento V, Souza VCd, Corado AL, Nascimento F, Silva G, Costa Á, Duarte D, Pessoa K, Mejía M, Brandão MJ, Jesus M, Gonçalves L, Costa CFd, Sampaio V, Barros D, Silva M, Mattos T, Pontes G, Abdalla L, Santos JH, Arantes I, Dezordi FZ, Siqueira MM, Wallau GL, Resende PC, Delatorre E, Gräf T, Bello G. 2021. COVID-19 in Amazonas, Brazil, was driven by the persistence of endemic lineages and P.1 emergence. Nat Med 27:1230–1238. doi:10.1038/s41591-021-01378-7. - DOI - PubMed
-
- Faria NR, Mellan TA, Whittaker C, Claro IM, Candido DDS, Mishra S, Crispim MAE, Sales FCS, Hawryluk I, McCrone JT, Hulswit RJG, Franco LAM, Ramundo MS, de Jesus JG, Andrade PS, Coletti TM, Ferreira GM, Silva CAM, Manuli ER, Pereira RHM, Peixoto PS, Kraemer MUG, Gaburo N, Jr, Camilo CDC, Hoeltgebaum H, Souza WM, Rocha EC, de Souza LM, de Pinho MC, Araujo LJT, Malta FSV, de Lima AB, Silva JDP, Zauli DAG, Ferreira ACS, Schnekenberg RP, Laydon DJ, Walker PGT, Schluter HM, Dos Santos ALP, Vidal MS, Del Caro VS, Filho RMF, Dos Santos HM, Aguiar RS, Proenca-Modena JL, Nelson B, Hay JA, Monod M, Miscouridou X, et al. . 2021. Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil. Science 372:815–821. doi:10.1126/science.abh2644. - DOI - PMC - PubMed
-
- Governo do Estado do Amazonas, Fundação de Vigilância em Saúde do Amazonas. 2021. Vacinômetro - COVID-19. https://www.fvs.am.gov.br/indicadorSalaSituacao_view/75/2. Accessed July 03, 2021.
Publication types
MeSH terms
Substances
Grants and funding
- LNCC
- E-26/202.896/2018/Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ)
- VPPCB-007-FIO-18-2-30/Fundação Oswaldo Cruz (FIOCRUZ)
- PCTI-EmergeSaude/Fundação de Amparo à Pesquisa do Estado do Amazonas (FAPEAM)
- Rede Genô/Fundação de Amparo à Pesquisa do Estado do Amazonas (FAPEAM)
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

