Mini-Review: Transgenerational CRISPR/Cas9 Gene Editing in Plants
- PMID: 35187531
- PMCID: PMC8854858
- DOI: 10.3389/fgeed.2022.825042
Mini-Review: Transgenerational CRISPR/Cas9 Gene Editing in Plants
Abstract
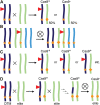
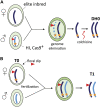
CRISPR/Cas9 genome editing has been used extensively in a wide variety of plant species. Creation of loss-of-function alleles, promoter variants and mutant collections are a few of the many uses of genome editing. In a typical workflow for sexually reproducing species, plants are generated that contain an integrated CRISPR/Cas9 transgene. After editing of the gene of interest, T-DNA null segregants can be identified in the next generation that contain only the desired edit. However, maintained presence of the CRISPR/Cas9 transgene and continued editing in the subsequent generations offer a range of applications for model plants and crops. In this review, we define transgenerational gene editing (TGE) as the continued editing of CRISPR/Cas9 after a genetic cross. We discuss the concept of TGE, summarize the current main applications, and highlight special cases to illustrate the importance of TGE for plant genome editing research and breeding.
Keywords: CRISPR/Cas9; HI-Edit; egg cell; floral dip; gene editing; pollen.
Copyright © 2022 Impens, Jacobs, Nelissen, Inzé and Pauwels.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
The present and potential future methods for delivering CRISPR/Cas9 components in plants.J Genet Eng Biotechnol. 2020 Jul 7;18(1):25. doi: 10.1186/s43141-020-00036-8. J Genet Eng Biotechnol. 2020. PMID: 32638190 Free PMC article. Review.
-
Precision genome editing in plants: state-of-the-art in CRISPR/Cas9-based genome engineering.BMC Plant Biol. 2020 May 25;20(1):234. doi: 10.1186/s12870-020-02385-5. BMC Plant Biol. 2020. PMID: 32450802 Free PMC article. Review.
-
The CRISPR/Cas9 system and its applications in crop genome editing.Crit Rev Biotechnol. 2019 May;39(3):321-336. doi: 10.1080/07388551.2018.1554621. Epub 2019 Jan 15. Crit Rev Biotechnol. 2019. PMID: 30646772 Review.
-
CRISPR/Cas9-mediated genome editing and gene replacement in plants: Transitioning from lab to field.Plant Sci. 2015 Nov;240:130-42. doi: 10.1016/j.plantsci.2015.09.011. Epub 2015 Sep 11. Plant Sci. 2015. PMID: 26475194 Review.
-
CRISPR/Cas9 genome editing through in planta transformation.Crit Rev Biotechnol. 2020 Mar;40(2):153-168. doi: 10.1080/07388551.2019.1709795. Epub 2020 Jan 5. Crit Rev Biotechnol. 2020. PMID: 31903793 Review.
Cited by
-
Breeding for improved digestibility and processing of lignocellulosic biomass in Zea mays.Front Plant Sci. 2024 Jul 26;15:1419796. doi: 10.3389/fpls.2024.1419796. eCollection 2024. Front Plant Sci. 2024. PMID: 39129761 Free PMC article. Review.
-
Usefulness of current sgRNA design guidelines and in vitro cleavage assays for plant CRISPR/Cas genome editing: a case targeting the polyphenol oxidase gene family in eggplant (Solanum melongena L.).Transgenic Res. 2023 Dec;32(6):561-573. doi: 10.1007/s11248-023-00371-9. Epub 2023 Oct 24. Transgenic Res. 2023. PMID: 37874448
-
Strategies and Methods for Improving the Efficiency of CRISPR/Cas9 Gene Editing in Plant Molecular Breeding.Plants (Basel). 2023 Mar 28;12(7):1478. doi: 10.3390/plants12071478. Plants (Basel). 2023. PMID: 37050104 Free PMC article. Review.
-
BREEDIT: a multiplex genome editing strategy to improve complex quantitative traits in maize.Plant Cell. 2023 Jan 2;35(1):218-238. doi: 10.1093/plcell/koac243. Plant Cell. 2023. PMID: 36066192 Free PMC article.
-
Current insights and advances into plant male sterility: new precision breeding technology based on genome editing applications.Front Plant Sci. 2023 Jul 13;14:1223861. doi: 10.3389/fpls.2023.1223861. eCollection 2023. Front Plant Sci. 2023. PMID: 37521915 Free PMC article. Review.
References
-
- Coe E. H. (1959). A Line of Maize with High Haploid Frequency. The Am. Naturalist 93, 381–382. 10.1086/282098 - DOI
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

