CCR2+ Macrophages Promote Orthodontic Tooth Movement and Alveolar Bone Remodeling
- PMID: 35185928
- PMCID: PMC8854866
- DOI: 10.3389/fimmu.2022.835986
CCR2+ Macrophages Promote Orthodontic Tooth Movement and Alveolar Bone Remodeling
Abstract
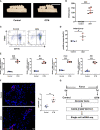
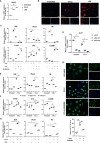
During mechanical force-induced alveolar bone remodeling, macrophage-mediated local inflammation plays a critical role. Yet, the detailed heterogeneity of macrophages is still unknown. Single-cell RNA sequencing was used to study the transcriptome heterogeneity of macrophages during alveolar bone remodeling. We identified macrophage subclusters with specific gene expression profiles and functions. CellChat and trajectory analysis revealed a central role of the Ccr2 cluster during development, with the CCL signaling pathway playing a crucial role. We further demonstrated that the Ccr2 cluster modulated bone remodeling associated inflammation through an NF-κB dependent pathway. Blocking CCR2 could significantly reduce the Orthodontic tooth movement (OTM) progression. In addition, we confirmed the variation of CCR2+ macrophages in human periodontal tissues. Our findings reveal that mechanical force-induced functional shift of the Ccr2 macrophages cluster mediated by NF-κB pathway, leading to a pro-inflammatory response and bone remodeling. This macrophage cluster may represent a potential target for the manipulation of OTM.
Keywords: CCR2; bone remodeling; inflammatory; macrophage; single-cell sequencing.
Copyright © 2022 Xu, Zhang, Sathe, Jin, Guan, Sun, Xing, Zhang and Yan.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






Similar articles
-
microRNA-21 Contributes to Orthodontic Tooth Movement.J Dent Res. 2016 Nov;95(12):1425-1433. doi: 10.1177/0022034516657043. Epub 2016 Jul 20. J Dent Res. 2016. PMID: 27422860
-
Role of CCR2 in orthodontic tooth movement.Am J Orthod Dentofacial Orthop. 2012 Feb;141(2):153-60. doi: 10.1016/j.ajodo.2011.07.019. Am J Orthod Dentofacial Orthop. 2012. PMID: 22284282
-
Lysozyme 1 Inflamed CCR2+ Macrophages Promote Obesity-Induced Cardiac Dysfunction.Circ Res. 2024 Aug 16;135(5):596-613. doi: 10.1161/CIRCRESAHA.124.324106. Epub 2024 Jul 26. Circ Res. 2024. PMID: 39056179
-
Immune Changes Induced by Orthodontic Forces: A Critical Review.J Dent Res. 2022 Jan;101(1):11-20. doi: 10.1177/00220345211016285. Epub 2021 Jun 9. J Dent Res. 2022. PMID: 34105404 Review.
-
Neural regulation of alveolar bone remodeling and periodontal ligament metabolism during orthodontic tooth movement in response to therapeutic loading.J World Fed Orthod. 2022 Oct;11(5):139-145. doi: 10.1016/j.ejwf.2022.08.003. Epub 2022 Sep 27. J World Fed Orthod. 2022. PMID: 36175332 Review.
Cited by
-
Mechanical force increases tooth movement and promotes remodeling of alveolar bone defects augmented with bovine bone mineral.Prog Orthod. 2024 Jan 8;25(1):2. doi: 10.1186/s40510-023-00501-3. Prog Orthod. 2024. PMID: 38185724 Free PMC article.
-
Identifying MS4A6A+ macrophages as potential contributors to the pathogenesis of nonalcoholic fatty liver disease, periodontitis, and type 2 diabetes mellitus.Heliyon. 2024 Apr 9;10(8):e29340. doi: 10.1016/j.heliyon.2024.e29340. eCollection 2024 Apr 30. Heliyon. 2024. PMID: 38644829 Free PMC article.
-
Effect of endodontically treated teeth on prosthetically guided orthodontics with clear aligners: a case series.BMC Oral Health. 2024 Oct 18;24(1):1242. doi: 10.1186/s12903-024-05007-w. BMC Oral Health. 2024. PMID: 39425114 Free PMC article.
-
Orthodontic Compression Enhances Macrophage M2 Polarization via Histone H3 Hyperacetylation.Int J Mol Sci. 2023 Feb 4;24(4):3117. doi: 10.3390/ijms24043117. Int J Mol Sci. 2023. PMID: 36834533 Free PMC article.
-
Mechanical force induces macrophage-derived exosomal UCHL3 promoting bone marrow mesenchymal stem cell osteogenesis by targeting SMAD1.J Nanobiotechnology. 2023 Mar 14;21(1):88. doi: 10.1186/s12951-023-01836-z. J Nanobiotechnology. 2023. PMID: 36915132 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

