METTL3 promotes oxaliplatin resistance of gastric cancer CD133+ stem cells by promoting PARP1 mRNA stability
- PMID: 35179655
- PMCID: PMC11072755
- DOI: 10.1007/s00018-022-04129-0
METTL3 promotes oxaliplatin resistance of gastric cancer CD133+ stem cells by promoting PARP1 mRNA stability
Abstract
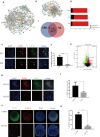
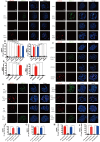
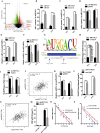
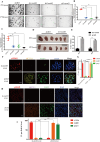
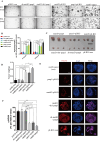
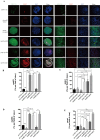
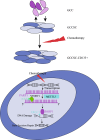
Oxaliplatin is the first-line regime for advanced gastric cancer treatment, while its resistance is a major problem that leads to the failure of clinical treatments. Tumor cell heterogeneity has been considered as one of the main causes for drug resistance in cancer. In this study, the mechanism of oxaliplatin resistance was investigated through in vitro human gastric cancer organoids and gastric cancer oxaliplatin-resistant cell lines and in vivo subcutaneous tumorigenicity experiments. The in vitro and in vivo results indicated that CD133+ stem cell-like cells are the main subpopulation and PARP1 is the central gene mediating oxaliplatin resistance in gastric cancer. It was found that PARP1 can effectively repair DNA damage caused by oxaliplatin by means of mediating the opening of base excision repair pathway, leading to the occurrence of drug resistance. The CD133+ stem cells also exhibited upregulated expression of N6-methyladenosine (m6A) mRNA and its writer METTL3 as showed by immunoprecipitation followed by sequencing and transcriptome analysis. METTTL3 enhances the stability of PARP1 by recruiting YTHDF1 to target the 3'-untranslated Region (3'-UTR) of PARP1 mRNA. The CD133+ tumor stem cells can regulate the stability and expression of m6A to PARP1 through METTL3, and thus exerting the PARP1-mediated DNA damage repair ability. Therefore, our study demonstrated that m6A Methyltransferase METTL3 facilitates oxaliplatin resistance in CD133+ gastric cancer stem cells by Promoting PARP1 mRNA stability which increases base excision repair pathway activity.
Keywords: Chemotherapy resistance; DNA repair; Digestive system tumors; Epigenetic modulation.
© 2022. The Author(s), under exclusive licence to Springer Nature Switzerland AG.
Conflict of interest statement
No potential conflicts of interest were disclosed.
Figures


Similar articles
-
Bone marrow-derived mesenchymal stem cells increase drug resistance in CD133-expressing gastric cancer cells by regulating the PI3K/AKT pathway.Tumour Biol. 2016 Nov;37(11):14637-14651. doi: 10.1007/s13277-016-5319-0. Epub 2016 Sep 12. Tumour Biol. 2016. PMID: 27619680
-
METTL3-mediated N6-methyladenosine modification is critical for epithelial-mesenchymal transition and metastasis of gastric cancer.Mol Cancer. 2019 Oct 13;18(1):142. doi: 10.1186/s12943-019-1065-4. Mol Cancer. 2019. PMID: 31607270 Free PMC article.
-
TOPBP1 regulates resistance of gastric cancer to oxaliplatin by promoting transcription of PARP1.DNA Repair (Amst). 2022 Mar;111:103278. doi: 10.1016/j.dnarep.2022.103278. Epub 2022 Feb 1. DNA Repair (Amst). 2022. PMID: 35124372
-
m6A mRNA methylation initiated by METTL3 directly promotes YAP translation and increases YAP activity by regulating the MALAT1-miR-1914-3p-YAP axis to induce NSCLC drug resistance and metastasis.J Hematol Oncol. 2019 Dec 9;12(1):135. doi: 10.1186/s13045-019-0830-6. J Hematol Oncol. 2019. Retraction in: J Hematol Oncol. 2023 Feb 22;16(1):14. doi: 10.1186/s13045-023-01416-6. PMID: 31818312 Free PMC article. Retracted.
-
Functions of methyltransferase-like 3 in breast cancer: pathogenesis, drug resistance, and therapeutic target.Breast Cancer Res. 2024 Jul 3;26(1):110. doi: 10.1186/s13058-024-01869-8. Breast Cancer Res. 2024. PMID: 38961497 Free PMC article. Review.
Cited by
-
GMEB2 Promotes the Growth of Colorectal Cancer by Activating ADRM1 Transcription and NF-κB Signalling and Is Positively Regulated by the m6A Reader YTHDF1.Cancers (Basel). 2022 Dec 8;14(24):6046. doi: 10.3390/cancers14246046. Cancers (Basel). 2022. PMID: 36551532 Free PMC article.
-
The Potential Value of m6A RNA Methylation in the Development of Cancers Focus on Malignant Glioma.Front Immunol. 2022 May 30;13:917153. doi: 10.3389/fimmu.2022.917153. eCollection 2022. Front Immunol. 2022. PMID: 35711459 Free PMC article. Review.
-
Role of RNA methylation in the regulation of pancreatic cancer stem cells (Review).Oncol Lett. 2023 Jun 20;26(2):336. doi: 10.3892/ol.2023.13922. eCollection 2023 Aug. Oncol Lett. 2023. PMID: 37427348 Free PMC article. Review.
-
Capecitabine enhances sensitivity to oxaliplatin in advanced gastric cancer and the effects on patients' FOXP1 and GGT levels.Heliyon. 2024 Oct 9;10(21):e39152. doi: 10.1016/j.heliyon.2024.e39152. eCollection 2024 Nov 15. Heliyon. 2024. PMID: 39524860 Free PMC article.
-
FOXA1/MND1/TKT axis regulates gastric cancer progression and oxaliplatin sensitivity via PI3K/AKT signaling pathway.Cancer Cell Int. 2023 Oct 10;23(1):234. doi: 10.1186/s12935-023-03077-4. Cancer Cell Int. 2023. PMID: 37817120 Free PMC article.
References
-
- Boku N, Ryu MH, Kato K, Chung HC, Minashi K, Lee KW, et al. Safety and efficacy of nivolumab in combination with S-1/capecitabine plus oxaliplatin in patients with previously untreated, unresectable, advanced, or recurrent gastric/gastroesophageal junction cancer: interim results of a randomized, phase II trial (ATTRACTION-4) Ann Oncol Off J Eur Soc Med Oncol. 2019;30(2):250–258. doi: 10.1093/annonc/mdy540. - DOI - PMC - PubMed
-
- Harada K, Sakamoto N, Ukai S, Yamamoto Y, Pham QT, Taniyama D, et al. Establishment of oxaliplatin-resistant gastric cancer organoids: importance of myoferlin in the acquisition of oxaliplatin resistance. Gastric Cancer 2021. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

