Considerations and practical implications of performing a phenotypic CRISPR/Cas survival screen
- PMID: 35176052
- PMCID: PMC8853573
- DOI: 10.1371/journal.pone.0263262
Considerations and practical implications of performing a phenotypic CRISPR/Cas survival screen
Abstract
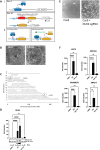
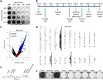
Genome-wide screens that have viability as a readout have been instrumental to identify essential genes. The development of gene knockout screens with the use of CRISPR-Cas has provided a more sensitive method to identify these genes. Here, we performed an exhaustive genome-wide CRISPR/Cas9 phenotypic rescue screen to identify modulators of cytotoxicity induced by the pioneer transcription factor, DUX4. Misexpression of DUX4 due to a failure in epigenetic repressive mechanisms underlies facioscapulohumeral muscular dystrophy (FHSD), a complex muscle disorder that thus far remains untreatable. As the name implies, FSHD generally starts in the muscles of the face and shoulder girdle. Our CRISPR/Cas9 screen revealed no key effectors other than DUX4 itself that could modulate DUX4 cytotoxicity, suggesting that treatment efforts in FSHD should be directed towards direct modulation of DUX4 itself. Our screen did however reveal some rare and unexpected genomic events, that had an important impact on the interpretation of our data. Our findings may provide important considerations for planning future CRISPR/Cas9 phenotypic survival screens.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Applying genome-wide CRISPR-Cas9 screens for therapeutic discovery in facioscapulohumeral muscular dystrophy.Sci Transl Med. 2020 Mar 25;12(536):eaay0271. doi: 10.1126/scitranslmed.aay0271. Sci Transl Med. 2020. PMID: 32213627 Free PMC article.
-
A patient-derived iPSC model revealed oxidative stress increases facioscapulohumeral muscular dystrophy-causative DUX4.Hum Mol Genet. 2018 Dec 1;27(23):4024-4035. doi: 10.1093/hmg/ddy293. Hum Mol Genet. 2018. PMID: 30107443 Free PMC article.
-
DUX4-induced constitutive DNA damage and oxidative stress contribute to aberrant differentiation of myoblasts from FSHD patients.Free Radic Biol Med. 2016 Oct;99:244-258. doi: 10.1016/j.freeradbiomed.2016.08.007. Epub 2016 Aug 9. Free Radic Biol Med. 2016. PMID: 27519269
-
The Genetics and Epigenetics of Facioscapulohumeral Muscular Dystrophy.Annu Rev Genomics Hum Genet. 2019 Aug 31;20:265-291. doi: 10.1146/annurev-genom-083118-014933. Epub 2019 Apr 24. Annu Rev Genomics Hum Genet. 2019. PMID: 31018108 Review.
-
Deciphering transcription dysregulation in FSH muscular dystrophy.J Hum Genet. 2012 Aug;57(8):477-84. doi: 10.1038/jhg.2012.74. Epub 2012 Jun 21. J Hum Genet. 2012. PMID: 22718021 Free PMC article. Review.
Cited by
-
Proteome-wide systems genetics identifies UFMylation as a regulator of skeletal muscle function.Elife. 2022 Dec 6;11:e82951. doi: 10.7554/eLife.82951. Elife. 2022. PMID: 36472367 Free PMC article.
-
Functional genetics reveals modulators of anti-microtubule drug sensitivity.bioRxiv [Preprint]. 2024 Mar 13:2024.03.12.584469. doi: 10.1101/2024.03.12.584469. bioRxiv. 2024. Update in: J Cell Biol. 2025 Feb 3;224(2):e202403065. doi: 10.1083/jcb.202403065. PMID: 38559203 Free PMC article. Updated. Preprint.
-
Efficient generation of lower induced motor neurons by coupling Ngn2 expression with developmental cues.Cell Rep. 2023 Jan 31;42(1):111896. doi: 10.1016/j.celrep.2022.111896. Epub 2023 Jan 2. Cell Rep. 2023. PMID: 36596304 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

