Uip4p modulates nuclear pore complex function in Saccharomyces cerevisiae
- PMID: 35171083
- PMCID: PMC8855845
- DOI: 10.1080/19491034.2022.2034286
Uip4p modulates nuclear pore complex function in Saccharomyces cerevisiae
Abstract
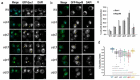
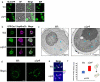
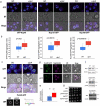
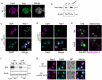
A double membrane bilayer perforated by nuclear pore complexes (NPCs) governs the shape of the nucleus, the prominent distinguishing organelle of a eukaryotic cell. Despite the absence of lamins in yeasts, the nuclear morphology is stably maintained and shape changes occur in a regulated fashion. In a quest to identify factors that contribute to regulation of nuclear shape and function in Saccharomyces cerevisiae, we used a fluorescence imaging based approach. Here we report the identification of a novel protein, Uip4p, that is required for regulation of nuclear morphology. Loss of Uip4 compromises NPC function and loss of nuclear envelope (NE) integrity. Our localization studies show that Uip4 localizes to the NE and endoplasmic reticulum (ER) network. Furthermore, we demonstrate that the localization and expression of Uip4 is regulated during growth, which is crucial for NPC distribution.
Keywords: Nuclear envelope; Uip4; nuclear pore complex; yeast.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures
Similar articles
-
The yeast integral membrane protein Apq12 potentially links membrane dynamics to assembly of nuclear pore complexes.J Cell Biol. 2007 Aug 27;178(5):799-812. doi: 10.1083/jcb.200702120. J Cell Biol. 2007. PMID: 17724120 Free PMC article.
-
Nuclear envelope insertion of spindle pole bodies and nuclear pore complexes.Nucleus. 2012 May-Jun;3(3):226-36. doi: 10.4161/nucl.20148. Epub 2012 May 1. Nucleus. 2012. PMID: 22572959 Free PMC article. Review.
-
NPC-phagy: selective autophagy of the nuclear pore complexes.Autophagy. 2020 Oct;16(10):1735-1736. doi: 10.1080/15548627.2020.1798199. Epub 2020 Jul 27. Autophagy. 2020. PMID: 32713250 Free PMC article.
-
Nuclear pore complex integrity requires Lnp1, a regulator of cortical endoplasmic reticulum.Mol Biol Cell. 2015 Aug 1;26(15):2833-44. doi: 10.1091/mbc.E15-01-0053. Epub 2015 Jun 3. Mol Biol Cell. 2015. PMID: 26041935 Free PMC article.
-
Nuclear pore complex biogenesis.Curr Opin Cell Biol. 2009 Aug;21(4):603-12. doi: 10.1016/j.ceb.2009.05.001. Epub 2009 Jun 11. Curr Opin Cell Biol. 2009. PMID: 19524430 Free PMC article. Review.
Cited by
-
An adaptable live-cell imaging protocol to analyze organelle morphology in Saccharomyces cerevisiae.STAR Protoc. 2022 Feb 14;3(1):101124. doi: 10.1016/j.xpro.2022.101124. eCollection 2022 Mar 18. STAR Protoc. 2022. PMID: 35243366 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
