N-Acetyltransferase 8 Promotes Viral Replication by Increasing the Stability of Enterovirus 71 Nonstructural Proteins
- PMID: 35170979
- PMCID: PMC8941898
- DOI: 10.1128/jvi.00119-22
N-Acetyltransferase 8 Promotes Viral Replication by Increasing the Stability of Enterovirus 71 Nonstructural Proteins
Erratum in
-
Erratum for Zhao et al., "N-Acetyltransferase 8 Promotes Viral Replication by Increasing the Stability of Enterovirus 71 Nonstructural Proteins".J Virol. 2023 Mar 30;97(3):e0025723. doi: 10.1128/jvi.00257-23. Epub 2023 Mar 14. J Virol. 2023. PMID: 36916908 Free PMC article. No abstract available.
Abstract
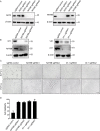
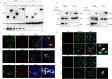
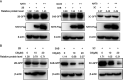
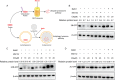
Enterovirus 71 (EV71) is deemed a reemergent pathogen, with recent outbreaks worldwide. EV71 infection causes hand, foot, and mouth disease (HFMD) and has been associated with severe cardiac and central nervous system complications and even death. Viruses need host factors to complete their life cycle; therefore, the identification of the host factors for EV71 infection is pivotal to new antiviral research. Emerging evidence has highlighted the importance of protein acetylation during infection by various human viruses. The endoplasmic reticulum (ER), as the prominent organelle of EV71 replication, also has a unique acetylation regulation mechanism. However, the pathogenesis of EV71 and its relationship with the ER-based acetylation machinery are not fully understood. In this study, we demonstrated for the first time that the ER-resident acetyltransferase N-acetyltransferase 8 (NAT8) is a host factor for EV71 infection. Inhibiting NAT8 with CRISPR or a small compound significantly suppressed EV71 infection in SK-N-SH cells. NAT8 promoted EV71 replication in an acetyltransferase-activity-dependent manner. Additionally, we found that NAT8 facilitates EV71 infection by interacting with EV71 2B, 3AB, and 3C proteins and increasing the stability of these proteins. These results uncovered a novel function of NAT8 and elucidated a new mechanism underlying the regulation of EV71 replication. IMPORTANCE EV71 is one of the most common pathogens causing HFMD in young children, and some patients experience severe or fatal neurological consequences. To ensure efficient replication, the virus must hijack multiple host factors for its own benefit. Here, we show that the ER-resident acetyltransferase NAT8 is a host factor for EV71 infection. EV71 fails to complete its infection in various cells in the absence of NAT8. We further show that NAT8 benefits EV71 replication in an acetyltransferase-activity-dependent manner. Finally, we show that NAT8 facilitates EV71 infection by interacting with EV71 2B, 3AB, and 3C proteins and increasing the stability of these proteins. These results uncovered a novel function of NAT8 in EV71 infection and elucidated a new mechanism underlying the regulation of EV71 replication.
Keywords: acetylation; enterovirus; viral replication.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
N-terminal acetyltransferase 6 facilitates enterovirus 71 replication by regulating PI4KB expression and replication organelle biogenesis.J Virol. 2024 Feb 20;98(2):e0174923. doi: 10.1128/jvi.01749-23. Epub 2024 Jan 8. J Virol. 2024. PMID: 38189249 Free PMC article.
-
The class III phosphatidylinositol 3-kinase VPS34 supports EV71 replication by promoting viral replication organelle formation.J Virol. 2024 Oct 22;98(10):e0069524. doi: 10.1128/jvi.00695-24. Epub 2024 Sep 10. J Virol. 2024. PMID: 39254312
-
Inhibition of enterovirus 71 replication and viral 3C protease by quercetin.Virol J. 2018 Jul 31;15(1):116. doi: 10.1186/s12985-018-1023-6. Virol J. 2018. PMID: 30064445 Free PMC article.
-
Recent Progress on Functional Genomics Research of Enterovirus 71.Virol Sin. 2019 Feb;34(1):9-21. doi: 10.1007/s12250-018-0071-9. Epub 2018 Dec 14. Virol Sin. 2019. PMID: 30552635 Free PMC article. Review.
-
Cell and tissue tropism of enterovirus 71 and other enteroviruses infections.J Biomed Sci. 2014 Mar 7;21(1):18. doi: 10.1186/1423-0127-21-18. J Biomed Sci. 2014. PMID: 24602216 Free PMC article. Review.
Cited by
-
MAVS-Based Reporter Systems for Real-Time Imaging of EV71 Infection and Antiviral Testing.Viruses. 2023 Apr 26;15(5):1064. doi: 10.3390/v15051064. Viruses. 2023. PMID: 37243150 Free PMC article.
-
N-terminal acetyltransferase 6 facilitates enterovirus 71 replication by regulating PI4KB expression and replication organelle biogenesis.J Virol. 2024 Feb 20;98(2):e0174923. doi: 10.1128/jvi.01749-23. Epub 2024 Jan 8. J Virol. 2024. PMID: 38189249 Free PMC article.
-
Cathelicidin peptide analogues inhibit EV71 infection through blocking viral entry and uncoating.PLoS Pathog. 2024 Jan 25;20(1):e1011967. doi: 10.1371/journal.ppat.1011967. eCollection 2024 Jan. PLoS Pathog. 2024. PMID: 38271479 Free PMC article.
-
Tribbles pseudokinase 3 promotes enterovirus A71 infection via dual mechanisms.Emerg Microbes Infect. 2024 Dec;13(1):2307514. doi: 10.1080/22221751.2024.2307514. Epub 2024 Jan 30. Emerg Microbes Infect. 2024. PMID: 38240287 Free PMC article.
-
N-Acetyltransferase 9 Inhibits Porcine Reproductive and Respiratory Syndrome Virus Proliferation by N-Terminal Acetylation of the Structural Protein GP5.Microbiol Spectr. 2023 Feb 14;11(1):e0244222. doi: 10.1128/spectrum.02442-22. Epub 2023 Jan 25. Microbiol Spectr. 2023. PMID: 36695606 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

