Hypoxia/Ischemia-Induced Rod Microglia Phenotype in CA1 Hippocampal Slices
- PMID: 35163344
- PMCID: PMC8836225
- DOI: 10.3390/ijms23031422
Hypoxia/Ischemia-Induced Rod Microglia Phenotype in CA1 Hippocampal Slices
Abstract
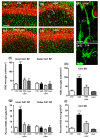
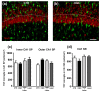
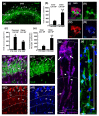
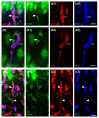
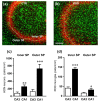
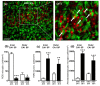
The complexity of microglia phenotypes and their related functions compels the continuous study of microglia in diseases animal models. We demonstrated that oxygen-glucose deprivation (OGD) induced rapid, time- and space-dependent phenotypic microglia modifications in CA1 stratum pyramidalis (SP) and stratum radiatum (SR) of rat organotypic hippocampal slices as well as the degeneration of pyramidal neurons, especially in the outer layer of SP. Twenty-four h following OGD, many rod microglia formed trains of elongated cells spanning from the SR throughout the CA1, reaching the SP outer layer where they acquired a round-shaped amoeboid phagocytic head and phagocytosed most of the pyknotic, damaged neurons. NIR-laser treatment, known to preserve neuronal viability after OGD, prevented rod microglia formation. In CA3 SP, pyramidal neurons were less damaged, no rod microglia were found. Thirty-six h after OGD, neuronal damage was more pronounced in SP outer and inner layers of CA1, rod microglia cells were no longer detectable, and most microglia were amoeboid/phagocytic. Damaged neurons, more numerous 36 h after OGD, were phagocytosed by amoeboid microglia in both inner and outer layers of CA1. In response to OGD, microglia can acquire different morphofunctional phenotypes which depend on the time after the insult and on the subregion where microglia are located.
Keywords: CA3 hippocampus; confocal microscopy; immunofluorescence; neurodegeneration; oxygen glucose deprivation; phagocytosis; rod microglia train.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Phenomic Microglia Diversity as a Druggable Target in the Hippocampus in Neurodegenerative Diseases.Int J Mol Sci. 2023 Sep 5;24(18):13668. doi: 10.3390/ijms241813668. Int J Mol Sci. 2023. PMID: 37761971 Free PMC article. Review.
-
Regional and time-dependent neuroprotective effect of hypothermia following oxygen-glucose deprivation.Hippocampus. 2015 Feb;25(2):197-207. doi: 10.1002/hipo.22364. Epub 2014 Sep 25. Hippocampus. 2015. PMID: 25212128
-
Neuroprotective effects of the anti-inflammatory compound triflusal on ischemia-like neurodegeneration in mouse hippocampal slice cultures occur independent of microglia.Exp Neurol. 2009 Jul;218(1):11-23. doi: 10.1016/j.expneurol.2009.03.023. Epub 2009 Mar 31. Exp Neurol. 2009. PMID: 19341733
-
The neuron-astrocyte-microglia triad in CA3 after chronic cerebral hypoperfusion in the rat: Protective effect of dipyridamole.Exp Gerontol. 2017 Oct 1;96:46-62. doi: 10.1016/j.exger.2017.06.006. Epub 2017 Jun 9. Exp Gerontol. 2017. PMID: 28606482
-
An Overview on the Differential Interplay Among Neurons-Astrocytes-Microglia in CA1 and CA3 Hippocampus in Hypoxia/Ischemia.Front Cell Neurosci. 2020 Nov 11;14:585833. doi: 10.3389/fncel.2020.585833. eCollection 2020. Front Cell Neurosci. 2020. PMID: 33262692 Free PMC article. Review.
Cited by
-
Modeling Central Nervous System Injury In Vitro: Current Status and Promising Future Strategies.Biomedicines. 2022 Dec 29;11(1):94. doi: 10.3390/biomedicines11010094. Biomedicines. 2022. PMID: 36672601 Free PMC article. Review.
-
Phenomic Microglia Diversity as a Druggable Target in the Hippocampus in Neurodegenerative Diseases.Int J Mol Sci. 2023 Sep 5;24(18):13668. doi: 10.3390/ijms241813668. Int J Mol Sci. 2023. PMID: 37761971 Free PMC article. Review.
-
Tau seeding and spreading in vivo is supported by both AD-derived fibrillar and oligomeric tau.Acta Neuropathol. 2023 Aug;146(2):191-210. doi: 10.1007/s00401-023-02600-1. Epub 2023 Jun 21. Acta Neuropathol. 2023. PMID: 37341831 Free PMC article.
-
The Protective Effect of CBD in a Model of In Vitro Ischemia May Be Mediated by Agonism on TRPV2 Channel and Microglia Activation.Int J Mol Sci. 2022 Oct 12;23(20):12144. doi: 10.3390/ijms232012144. Int J Mol Sci. 2022. PMID: 36292998 Free PMC article.
-
Microglia dynamic response and phenotype heterogeneity in neural regeneration following hypoxic-ischemic brain injury.Front Immunol. 2023 Nov 29;14:1320271. doi: 10.3389/fimmu.2023.1320271. eCollection 2023. Front Immunol. 2023. PMID: 38094292 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

