Flexible TALEs for an expanded use in gene activation, virulence and scaffold engineering
- PMID: 35150566
- PMCID: PMC8887545
- DOI: 10.1093/nar/gkac098
Flexible TALEs for an expanded use in gene activation, virulence and scaffold engineering
Abstract
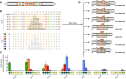
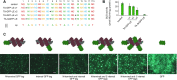
Transcription activator-like effectors (TALEs) are bacterial proteins with a programmable DNA-binding domain, which turned them into exceptional tools for biotechnology. TALEs contain a central array of consecutive 34 amino acid long repeats to bind DNA in a simple one-repeat-to-one-nucleotide manner. However, a few naturally occurring aberrant repeat variants break this strict binding mechanism, allowing for the recognition of an additional sequence with a -1 nucleotide frameshift. The limits and implications of this extended TALE binding mode are largely unexplored. Here, we analyse the complete diversity of natural and artificially engineered aberrant repeats for their impact on the DNA binding of TALEs. Surprisingly, TALEs with several aberrant repeats can loop out multiple repeats simultaneously without losing DNA-binding capacity. We also characterized members of the only natural TALE class harbouring two aberrant repeats and confirmed that their target is the major virulence factor OsSWEET13 from rice. In an aberrant TALE repeat, the position and nature of the amino acid sequence strongly influence its function. We explored the tolerance of TALE repeats towards alterations further and demonstrate that inserts as large as GFP can be tolerated without disrupting DNA binding. This illustrates the extraordinary DNA-binding capacity of TALEs and opens new uses in biotechnology.
© The Author(s) 2022. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




Similar articles
-
Evolution of Transcription Activator-Like Effectors in Xanthomonas oryzae.Genome Biol Evol. 2017 Jun 1;9(6):1599-1615. doi: 10.1093/gbe/evx108. Genome Biol Evol. 2017. PMID: 28637323 Free PMC article.
-
A TAL effector repeat architecture for frameshift binding.Nat Commun. 2014 Mar 11;5:3447. doi: 10.1038/ncomms4447. Nat Commun. 2014. PMID: 24614980
-
Engineered TALE Repeats for Enhanced Imaging-Based Analysis of Cellular 5-Methylcytosine.Chembiochem. 2021 Feb 15;22(4):645-651. doi: 10.1002/cbic.202000563. Epub 2020 Nov 6. Chembiochem. 2021. PMID: 32991020 Free PMC article.
-
TALE: a tale of genome editing.Prog Biophys Mol Biol. 2014 Jan;114(1):25-32. doi: 10.1016/j.pbiomolbio.2013.11.006. Epub 2013 Nov 27. Prog Biophys Mol Biol. 2014. PMID: 24291598 Review.
-
[Advances in transcription activator-like effectors--a review].Sheng Wu Gong Cheng Xue Bao. 2015 Jul;31(7):1024-38. Sheng Wu Gong Cheng Xue Bao. 2015. PMID: 26647578 Review. Chinese.
Cited by
-
Designer TALEs enable discovery of cell death-inducer genes.Plant Physiol. 2024 Jul 31;195(4):2985-2996. doi: 10.1093/plphys/kiae230. Plant Physiol. 2024. PMID: 38723194 Free PMC article.
-
Insights into the Transcriptomics of Crop Wild Relatives to Unravel the Salinity Stress Adaptive Mechanisms.Int J Mol Sci. 2023 Jun 6;24(12):9813. doi: 10.3390/ijms24129813. Int J Mol Sci. 2023. PMID: 37372961 Free PMC article. Review.
References
-
- Boch J., Bonas U.. Xanthomonas AvrBs3 family-type III effectors: discovery and function. Annu. Rev. Phytopathol. 2010; 48:419–436. - PubMed
-
- Perez-Quintero A.L., Szurek B.. A decade decoded: spies and hackers in the history of TAL effectors research. Annu. Rev. Phytopathol. 2019; 57:459–481. - PubMed
-
- Wilkins K., Booher N., Wang L., Bogdanove A.. TAL effectors and activation of predicted host targets distinguish Asian from African strains of the rice pathogen Xanthomonasoryzae pv. oryzicola while strict conservation suggests universal importance of five TAL effectors. Front. Plant Sci. 2015; 6:536. - PMC - PubMed
-
- Tran T.T., Pérez-Quintero A.L., Wonni I., Carpenter S.C.D., Yu Y., Wang L., Leach J.E., Verdier V., Cunnac S., Bogdanove A.J.et al. .. Functional analysis of African Xanthomonasoryzae pv. oryzae TALomes reveals a new susceptibility gene in bacterial leaf blight of rice. PLoS Pathog. 2018; 14:e1007092. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

