Folding of VemP into translation-arresting secondary structure is driven by the ribosome exit tunnel
- PMID: 35150281
- PMCID: PMC8887479
- DOI: 10.1093/nar/gkac038
Folding of VemP into translation-arresting secondary structure is driven by the ribosome exit tunnel
Abstract
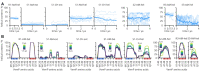
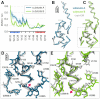
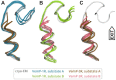
The ribosome is a fundamental biomolecular complex that synthesizes proteins in cells. Nascent proteins emerge from the ribosome through a tunnel, where they may interact with the tunnel walls or small molecules such as antibiotics. These interactions can cause translational arrest with notable physiological consequences. Here, we studied the arrest caused by the regulatory peptide VemP, which is known to form α-helices inside the ribosome tunnel near the peptidyl transferase center under specific conditions. We used all-atom molecular dynamics simulations of the entire ribosome and circular dichroism spectroscopy to study the driving forces of helix formation and how VemP causes the translational arrest. To that aim, we compared VemP dynamics in the ribosome tunnel with its dynamics in solution. We show that the VemP peptide has a low helical propensity in water and that the propensity is higher in mixtures of water and trifluorethanol. We propose that helix formation within the ribosome is driven by the interactions of VemP with the tunnel and that a part of VemP acts as an anchor. This anchor might slow down VemP progression through the tunnel enabling α-helix formation, which causes the elongation arrest.
© The Author(s) 2022. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

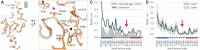
Similar articles
-
How the ribosome shapes cotranslational protein folding.Curr Opin Struct Biol. 2024 Feb;84:102740. doi: 10.1016/j.sbi.2023.102740. Epub 2023 Dec 9. Curr Opin Struct Biol. 2024. PMID: 38071940 Review.
-
The force-sensing peptide VemP employs extreme compaction and secondary structure formation to induce ribosomal stalling.Elife. 2017 May 30;6:e25642. doi: 10.7554/eLife.25642. Elife. 2017. PMID: 28556777 Free PMC article.
-
A switch from α-helical to β-strand conformation during co-translational protein folding.EMBO J. 2022 Feb 15;41(4):e109175. doi: 10.15252/embj.2021109175. Epub 2022 Jan 7. EMBO J. 2022. PMID: 34994471 Free PMC article.
-
Determinants of Helix Formation for a Kv1.3 Transmembrane Segment inside the Ribosome Exit Tunnel.J Mol Biol. 2017 Jun 2;429(11):1722-1732. doi: 10.1016/j.jmb.2017.04.022. Epub 2017 May 4. J Mol Biol. 2017. PMID: 28478285 Free PMC article.
-
Cotranslational folding--omnia mea mecum porto?Int J Biochem Cell Biol. 2001 Jun;33(6):541-53. doi: 10.1016/s1357-2725(01)00044-9. Int J Biochem Cell Biol. 2001. PMID: 11378437 Review.
Cited by
-
Ribosome Tunnel Environment Drives the Formation of α-Helix during Cotranslational Folding.J Chem Inf Model. 2024 Aug 26;64(16):6610-6622. doi: 10.1021/acs.jcim.4c00901. Epub 2024 Aug 16. J Chem Inf Model. 2024. PMID: 39150098 Free PMC article.
-
Probing Interplays between Human XBP1u Translational Arrest Peptide and 80S Ribosome.J Chem Theory Comput. 2022 Mar 8;18(3):1905-1914. doi: 10.1021/acs.jctc.1c00796. Epub 2021 Dec 9. J Chem Theory Comput. 2022. PMID: 34881571 Free PMC article.
-
Geometric differences in the ribosome exit tunnel impact the escape of small nascent proteins.Biophys J. 2023 Jan 3;122(1):20-29. doi: 10.1016/j.bpj.2022.11.2945. Epub 2022 Dec 5. Biophys J. 2023. PMID: 36463403 Free PMC article.
-
A nascent peptide code for translational control of mRNA stability in human cells.Nat Commun. 2022 Nov 11;13(1):6829. doi: 10.1038/s41467-022-34664-0. Nat Commun. 2022. PMID: 36369503 Free PMC article.
-
Binding of the peptide deformylase on the ribosome surface modulates the exit tunnel interior.Biophys J. 2022 Dec 6;121(23):4443-4451. doi: 10.1016/j.bpj.2022.11.004. Epub 2022 Nov 5. Biophys J. 2022. PMID: 36335428 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

