Determining if T cell antigens are naturally processed and presented on HLA class I molecules
- PMID: 35148673
- PMCID: PMC8832792
- DOI: 10.1186/s12865-022-00478-4
Determining if T cell antigens are naturally processed and presented on HLA class I molecules
Abstract
Background: Determining T cell responses to naturally processed and presented antigens is a critical immune correlate to determine efficacy of an investigational immunotherapeutic in clinical trials. In most cases, minimal epitopes and HLA restriction elements are unknown.
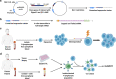
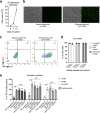
Results: Here, we detail the experimental use of ex vivo expanded autologous B cells as antigen presenting cells to overcome the limitation of unknown HLA restriction, and the use of electroporated full length mRNA encoding full length parental proteins to ensure that any observed T cell responses are specific for antigens that are naturally processed and presented.
Conclusions: This technique can serve as useful experimental approach to determine the induction or enhancement of specific responses to naturally processed and presented antigens on HLA class I molecules in peripheral blood or tumor infiltrating T cells.
Keywords: Antigen presenting cells; Electroporation; HLA restriction; Minimal epitope; T cell responses.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that no competing interests exist.
Figures

Similar articles
-
An in vitro-transcribed-mRNA polyepitope construct encoding 32 distinct HLA class I-restricted epitopes from CMV, EBV, and Influenza for use as a functional control in human immune monitoring studies.J Immunol Methods. 2010 Aug 31;360(1-2):149-56. doi: 10.1016/j.jim.2010.07.003. Epub 2010 Jul 15. J Immunol Methods. 2010. PMID: 20637775
-
Efficient presentation of known HLA class II-restricted MAGE-A3 epitopes by dendritic cells electroporated with messenger RNA encoding an invariant chain with genetic exchange of class II-associated invariant chain peptide.Cancer Res. 2003 Sep 1;63(17):5587-94. Cancer Res. 2003. PMID: 14500399
-
Melanoma-specific CD4+ T lymphocytes recognize human melanoma antigens processed and presented by Epstein-Barr virus-transformed B cells.Int J Cancer. 1994 Jul 1;58(1):69-79. doi: 10.1002/ijc.2910580113. Int J Cancer. 1994. PMID: 7516926
-
Identification of a naturally processed NY-ESO-1 peptide recognized by CD8+ T cells in the context of HLA-B51.Cancer Immun. 2002 Sep 19;2:12. Cancer Immun. 2002. PMID: 12747757
-
Antigen-presenting cells and the selection of immunodominant epitopes.Crit Rev Immunol. 1997;17(5-6):411-7. Crit Rev Immunol. 1997. PMID: 9419428 Review.
Cited by
-
Rapid identification of CMV-specific TCRs via reverse TCR cloning system based on bulk TCR repertoire data.Front Immunol. 2022 Nov 18;13:1021067. doi: 10.3389/fimmu.2022.1021067. eCollection 2022. Front Immunol. 2022. PMID: 36466875 Free PMC article.
-
Durable response in a patient with recurrent respiratory papillomatosis treated with immune checkpoint blockade.Head Neck. 2022 Oct;44(10):E31-E37. doi: 10.1002/hed.27144. Epub 2022 Jul 11. Head Neck. 2022. PMID: 35815785 Free PMC article.
-
Utilizing immunogenomic approaches to prioritize targetable neoantigens for personalized cancer immunotherapy.Front Immunol. 2023 Dec 12;14:1301100. doi: 10.3389/fimmu.2023.1301100. eCollection 2023. Front Immunol. 2023. PMID: 38149253 Free PMC article. Review.
-
Enhanced neoepitope-specific immunity following neoadjuvant PD-L1 and TGF-β blockade in HPV-unrelated head and neck cancer.J Clin Invest. 2022 Sep 15;132(18):e161400. doi: 10.1172/JCI161400. J Clin Invest. 2022. PMID: 35727629 Free PMC article. Clinical Trial.
References
-
- Burtness B, Harrington KJ, Greil R, Soulieres D, Tahara M, de Castro G, Jr, , et al. Pembrolizumab alone or with chemotherapy versus cetuximab with chemotherapy for recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-048): a randomised, open-label, phase 3 study. Lancet. 2019;394(10212):1915–1928. doi: 10.1016/S0140-6736(19)32591-7. - DOI - PubMed
-
- Massarelli E, William W, Johnson F, Kies M, Ferrarotto R, Guo M, et al. Combining immune checkpoint blockade and tumor-specific vaccine for patients with incurable human papillomavirus 16-related cancer: a phase 2 clinical trial. JAMA Oncol. 2019;5(1):67–73. doi: 10.1001/jamaoncol.2018.4051. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

