The actin nucleator Spir-1 is a virus restriction factor that promotes innate immune signalling
- PMID: 35148361
- PMCID: PMC8870497
- DOI: 10.1371/journal.ppat.1010277
The actin nucleator Spir-1 is a virus restriction factor that promotes innate immune signalling
Abstract
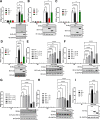
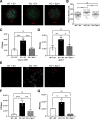
Cellular proteins often have multiple and diverse functions. This is illustrated with protein Spir-1 that is an actin nucleator, but, as shown here, also functions to enhance innate immune signalling downstream of RNA sensing by RIG-I/MDA-5. In human and mouse cells lacking Spir-1, IRF3 and NF-κB-dependent gene activation is impaired, whereas Spir-1 overexpression enhanced IRF3 activation. Furthermore, the infectious virus titres and sizes of plaques formed by two viruses that are sensed by RIG-I, vaccinia virus (VACV) and Zika virus, are increased in Spir-1 KO cells. These observations demonstrate the biological importance of Spir-1 in the response to virus infection. Like cellular proteins, viral proteins also have multiple and diverse functions. Here, we also show that VACV virulence factor K7 binds directly to Spir-1 and that a diphenylalanine motif of Spir-1 is needed for this interaction and for Spir-1-mediated enhancement of IRF3 activation. Thus, Spir-1 is a new virus restriction factor and is targeted directly by an immunomodulatory viral protein that enhances virus virulence and diminishes the host antiviral responses.
Conflict of interest statement
The authors have declared that no competing interests exist
Figures





Similar articles
-
DDX50 Is a Viral Restriction Factor That Enhances IRF3 Activation.Viruses. 2022 Feb 3;14(2):316. doi: 10.3390/v14020316. Viruses. 2022. PMID: 35215908 Free PMC article.
-
Vaccinia Virus BBK E3 Ligase Adaptor A55 Targets Importin-Dependent NF-κB Activation and Inhibits CD8+ T-Cell Memory.J Virol. 2019 May 1;93(10):e00051-19. doi: 10.1128/JVI.00051-19. Print 2019 May 15. J Virol. 2019. PMID: 30814284 Free PMC article.
-
Deletion of the K1L Gene Results in a Vaccinia Virus That Is Less Pathogenic Due to Muted Innate Immune Responses, yet Still Elicits Protective Immunity.J Virol. 2017 Jul 12;91(15):e00542-17. doi: 10.1128/JVI.00542-17. Print 2017 Aug 1. J Virol. 2017. PMID: 28490586 Free PMC article.
-
Structural and functional insights into the Spir/formin actin nucleator complex.Biol Chem. 2013 Dec;394(12):1649-60. doi: 10.1515/hsz-2013-0176. Biol Chem. 2013. PMID: 23863697 Review.
-
Vaccinia virus immune evasion: mechanisms, virulence and immunogenicity.J Gen Virol. 2013 Nov;94(Pt 11):2367-2392. doi: 10.1099/vir.0.055921-0. Epub 2013 Sep 2. J Gen Virol. 2013. PMID: 23999164 Review.
Cited by
-
The Viral Protein K7 Inhibits Biochemical Activities and Condensate Formation by the DEAD-box Helicase DDX3X.J Mol Biol. 2023 Oct 1;435(19):168217. doi: 10.1016/j.jmb.2023.168217. Epub 2023 Jul 28. J Mol Biol. 2023. PMID: 37517790 Free PMC article.
-
Exploiting RIG-I-like receptor pathway for cancer immunotherapy.J Hematol Oncol. 2023 Feb 8;16(1):8. doi: 10.1186/s13045-023-01405-9. J Hematol Oncol. 2023. PMID: 36755342 Free PMC article. Review.
-
Comparative genomics and integrated system biology approach unveiled undirected phylogeny patterns, mutational hotspots, functional patterns, and molecule repurposing for monkeypox virus.Funct Integr Genomics. 2023 Jul 11;23(3):231. doi: 10.1007/s10142-023-01168-z. Funct Integr Genomics. 2023. PMID: 37432480
-
Monkeypox virus: phylogenomics, host-pathogen interactome and mutational cascade.Microb Genom. 2023 Apr;9(4):mgen000987. doi: 10.1099/mgen.0.000987. Microb Genom. 2023. PMID: 37043267 Free PMC article.
-
The nuclear localization signal of monkeypox virus protein P2 orthologue is critical for inhibition of IRF3-mediated innate immunity.Emerg Microbes Infect. 2024 Dec;13(1):2372344. doi: 10.1080/22221751.2024.2372344. Epub 2024 Jul 6. Emerg Microbes Infect. 2024. PMID: 38916407 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

