Analysis of Pathogenic Pseudoexons Reveals Novel Mechanisms Driving Cryptic Splicing
- PMID: 35140743
- PMCID: PMC8819188
- DOI: 10.3389/fgene.2021.806946
Analysis of Pathogenic Pseudoexons Reveals Novel Mechanisms Driving Cryptic Splicing
Erratum in
-
Corrigendum: Analysis of Pathogenic Pseudoexons Reveals Novel Mechanisms Driving Cryptic Splicing.Front Genet. 2022 Jun 9;13:943044. doi: 10.3389/fgene.2022.943044. eCollection 2022. Front Genet. 2022. PMID: 35754842 Free PMC article.
Abstract
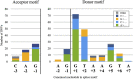
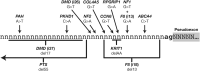
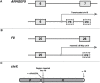
Understanding pre-mRNA splicing is crucial to accurately diagnosing and treating genetic diseases. However, mutations that alter splicing can exert highly diverse effects. Of all the known types of splicing mutations, perhaps the rarest and most difficult to predict are those that activate pseudoexons, sometimes also called cryptic exons. Unlike other splicing mutations that either destroy or redirect existing splice events, pseudoexon mutations appear to create entirely new exons within introns. Since exon definition in vertebrates requires coordinated arrangements of numerous RNA motifs, one might expect that pseudoexons would only arise when rearrangements of intronic DNA create novel exons by chance. Surprisingly, although such mutations do occur, a far more common cause of pseudoexons is deep-intronic single nucleotide variants, raising the question of why these latent exon-like tracts near the mutation sites have not already been purged from the genome by the evolutionary advantage of more efficient splicing. Possible answers may lie in deep intronic splicing processes such as recursive splicing or poison exon splicing. Because these processes utilize intronic motifs that benignly engage with the spliceosome, the regions involved may be more susceptible to exonization than other intronic regions would be. We speculated that a comprehensive study of reported pseudoexons might detect alignments with known deep intronic splice sites and could also permit the characterisation of novel pseudoexon categories. In this report, we present and analyse a catalogue of over 400 published pseudoexon splice events. In addition to confirming prior observations of the most common pseudoexon mutation types, the size of this catalogue also enabled us to suggest new categories for some of the rarer types of pseudoexon mutation. By comparing our catalogue against published datasets of non-canonical splice events, we also found that 15.7% of pseudoexons exhibit some splicing activity at one or both of their splice sites in non-mutant cells. Importantly, this included seven examples of experimentally confirmed recursive splice sites, confirming for the first time a long-suspected link between these two splicing phenomena. These findings have the potential to improve the fidelity of genetic diagnostics and reveal new targets for splice-modulating therapies.
Keywords: cryptic splicing; genetic disease; poison exons; pseudoexons; recursive splicing; splicing mutations.
Copyright © 2022 Keegan, Wilton and Fletcher.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Antisense Oligonucleotide Rescue of Deep-Intronic Variants Activating Pseudoexons in the 6-Pyruvoyl-Tetrahydropterin Synthase Gene.Nucleic Acid Ther. 2022 Oct;32(5):378-390. doi: 10.1089/nat.2021.0066. Epub 2022 Jul 12. Nucleic Acid Ther. 2022. PMID: 35833796 Free PMC article.
-
Pseudoexon activation in disease by non-splice site deep intronic sequence variation - wild type pseudoexons constitute high-risk sites in the human genome.Hum Mutat. 2022 Feb;43(2):103-127. doi: 10.1002/humu.24306. Epub 2021 Dec 5. Hum Mutat. 2022. PMID: 34837434 Review.
-
Silencer elements as possible inhibitors of pseudoexon splicing.Nucleic Acids Res. 2004 Mar 19;32(5):1783-91. doi: 10.1093/nar/gkh341. Print 2004. Nucleic Acids Res. 2004. PMID: 15034146 Free PMC article.
-
Multiple splicing defects in an intronic false exon.Mol Cell Biol. 2000 Sep;20(17):6414-25. doi: 10.1128/MCB.20.17.6414-6425.2000. Mol Cell Biol. 2000. PMID: 10938119 Free PMC article.
-
Splicing mutations in inherited retinal diseases.Prog Retin Eye Res. 2021 Jan;80:100874. doi: 10.1016/j.preteyeres.2020.100874. Epub 2020 Jun 15. Prog Retin Eye Res. 2021. PMID: 32553897 Review.
Cited by
-
Antisense Oligonucleotide Rescue of Deep-Intronic Variants Activating Pseudoexons in the 6-Pyruvoyl-Tetrahydropterin Synthase Gene.Nucleic Acid Ther. 2022 Oct;32(5):378-390. doi: 10.1089/nat.2021.0066. Epub 2022 Jul 12. Nucleic Acid Ther. 2022. PMID: 35833796 Free PMC article.
-
Prevalence, parameters, and pathogenic mechanisms for splice-altering acceptor variants that disrupt the AG exclusion zone.HGG Adv. 2022 Jun 25;3(4):100125. doi: 10.1016/j.xhgg.2022.100125. eCollection 2022 Oct 13. HGG Adv. 2022. PMID: 35847480 Free PMC article.
-
Age-Related Alternative Splicing: Driver or Passenger in the Aging Process?Cells. 2023 Dec 12;12(24):2819. doi: 10.3390/cells12242819. Cells. 2023. PMID: 38132139 Free PMC article. Review.
-
PDIVAS: Pathogenicity predictor for Deep-Intronic Variants causing Aberrant Splicing.BMC Genomics. 2023 Oct 10;24(1):601. doi: 10.1186/s12864-023-09645-2. BMC Genomics. 2023. PMID: 37817060 Free PMC article.
-
Updating mRNA variants of the human RSK4 gene and their expression in different stressed situations.Heliyon. 2024 Mar 8;10(7):e27475. doi: 10.1016/j.heliyon.2024.e27475. eCollection 2024 Apr 15. Heliyon. 2024. PMID: 38560189 Free PMC article.
References
-
- Ajeawung N. F., Nguyen T. T. M., Lu L., Kucharski T. J., Rousseau J., Molidperee S., et al. (2019). Mutations in ANAPC1, Encoding a Scaffold Subunit of the Anaphase-Promoting Complex, Cause Rothmund-Thomson Syndrome Type 1. Am. J. Hum. Genet. 105 (3), 625–630. 10.1016/j.ajhg.2019.06.011 - DOI - PMC - PubMed
-
- Albert S., Garanto A., Sangermano R., Khan M., Bax N. M., Hoyng C. B., et al. (2018). Identification and Rescue of Splice Defects Caused by Two Neighboring Deep-Intronic ABCA4 Mutations Underlying Stargardt Disease. Am. J. Hum. Genet. 102 (4), 517–527. 10.1016/j.ajhg.2018.02.008 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

