Epitranscriptomic regulation of HIV-1 full-length RNA packaging
- PMID: 35137199
- PMCID: PMC8887480
- DOI: 10.1093/nar/gkac062
Epitranscriptomic regulation of HIV-1 full-length RNA packaging
Erratum in
-
Correction to 'Epitranscriptomic regulation of HIV-1 full-length RNA packaging'.Nucleic Acids Res. 2022 May 6;50(8):4799. doi: 10.1093/nar/gkac281. Nucleic Acids Res. 2022. PMID: 35438791 Free PMC article. No abstract available.
Abstract
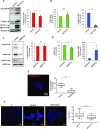
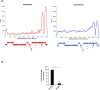
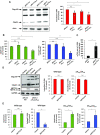
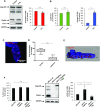
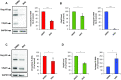
During retroviral replication, the full-length RNA serves both as mRNA and genomic RNA. However, the mechanisms by which the HIV-1 Gag protein selects the two RNA molecules that will be packaged into nascent virions remain poorly understood. Here, we demonstrate that deposition of N6-methyladenosine (m6A) regulates full-length RNA packaging. While m6A deposition by METTL3/METTL14 onto the full-length RNA was associated with increased Gag synthesis and reduced packaging, FTO-mediated demethylation promoted the incorporation of the full-length RNA into viral particles. Interestingly, HIV-1 Gag associates with the RNA demethylase FTO in the nucleus and contributes to full-length RNA demethylation. We further identified two highly conserved adenosines within the 5'-UTR that have a crucial functional role in m6A methylation and packaging of the full-length RNA. Together, our data propose a novel epitranscriptomic mechanism allowing the selection of the HIV-1 full-length RNA molecules that will be used as viral genomes.
© The Author(s) 2022. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
Similar articles
-
The Functions of N-methyladenosine (m6A) Modification on HIV-1 mRNA.Cell Biochem Biophys. 2024 Jun;82(2):561-574. doi: 10.1007/s12013-024-01280-2. Epub 2024 May 16. Cell Biochem Biophys. 2024. PMID: 38753251 Review.
-
Unpaired Guanosines in the 5' Untranslated Region of HIV-1 RNA Act Synergistically To Mediate Genome Packaging.J Virol. 2020 Oct 14;94(21):e00439-20. doi: 10.1128/JVI.00439-20. Print 2020 Oct 14. J Virol. 2020. PMID: 32796062 Free PMC article.
-
Interactions between HIV-1 Gag and Viral RNA Genome Enhance Virion Assembly.J Virol. 2017 Jul 27;91(16):e02319-16. doi: 10.1128/JVI.02319-16. Print 2017 Aug 15. J Virol. 2017. PMID: 28539452 Free PMC article.
-
Subcellular Localization of HIV-1 gag-pol mRNAs Regulates Sites of Virion Assembly.J Virol. 2017 Feb 28;91(6):e02315-16. doi: 10.1128/JVI.02315-16. Print 2017 Mar 15. J Virol. 2017. PMID: 28053097 Free PMC article.
-
Life of psi: how full-length HIV-1 RNAs become packaged genomes in the viral particles.Virology. 2014 Apr;454-455:362-70. doi: 10.1016/j.virol.2014.01.019. Epub 2014 Feb 14. Virology. 2014. PMID: 24530126 Free PMC article. Review.
Cited by
-
Recent insights into N6-methyladenosine during viral infection.Curr Opin Genet Dev. 2024 Aug;87:102213. doi: 10.1016/j.gde.2024.102213. Epub 2024 Jun 19. Curr Opin Genet Dev. 2024. PMID: 38901100 Review.
-
Comparative analysis of retroviral Gag-host cell interactions: focus on the nuclear interactome.bioRxiv [Preprint]. 2024 Mar 6:2024.01.18.575255. doi: 10.1101/2024.01.18.575255. bioRxiv. 2024. Update in: Retrovirology. 2024 Jun 19;21(1):13. doi: 10.1186/s12977-024-00645-y PMID: 38293010 Free PMC article. Updated. Preprint.
-
WTAP Targets the METTL3 m6A-Methyltransferase Complex to Cytoplasmic Hepatitis C Virus RNA to Regulate Infection.J Virol. 2022 Nov 23;96(22):e0099722. doi: 10.1128/jvi.00997-22. Epub 2022 Oct 31. J Virol. 2022. PMID: 36314819 Free PMC article.
-
The Functions of N-methyladenosine (m6A) Modification on HIV-1 mRNA.Cell Biochem Biophys. 2024 Jun;82(2):561-574. doi: 10.1007/s12013-024-01280-2. Epub 2024 May 16. Cell Biochem Biophys. 2024. PMID: 38753251 Review.
-
Mapping m6A Sites on HIV-1 RNA Using Oligonucleotide LC-MS/MS.Methods Protoc. 2024 Jan 10;7(1):7. doi: 10.3390/mps7010007. Methods Protoc. 2024. PMID: 38251200 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

