Increased mitochondrial proline metabolism sustains proliferation and survival of colorectal cancer cells
- PMID: 35130302
- PMCID: PMC8820619
- DOI: 10.1371/journal.pone.0262364
Increased mitochondrial proline metabolism sustains proliferation and survival of colorectal cancer cells
Abstract
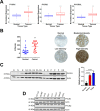
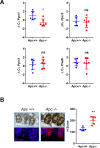
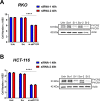
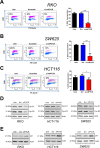
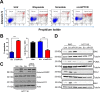
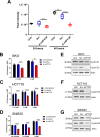
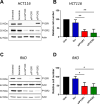
Research into the metabolism of the non-essential amino acid (NEAA) proline in cancer has gained traction in recent years. The last step in the proline biosynthesis pathway is catalyzed by pyrroline-5-carboxylate reductase (PYCR) enzymes. There are three PYCR enzymes: mitochondrial PYCR1 and 2 and cytosolic PYCR3 encoded by separate genes. The expression of the PYCR1 gene is increased in numerous malignancies and correlates with poor prognosis. PYCR1 expression sustains cancer cells' proliferation and survival and several mechanisms have been implicated to explain its oncogenic role. It has been suggested that the biosynthesis of proline is key to sustain protein synthesis, support mitochondrial function and nucleotide biosynthesis. However, the links between proline metabolism and cancer remain ill-defined and are likely to be tissue specific. Here we use a combination of human dataset, human tissue and mouse models to show that the expression levels of the proline biosynthesis enzymes are significantly increased during colorectal tumorigenesis. Functionally, the expression of mitochondrial PYCRs is necessary for cancer cells' survival and proliferation. However, the phenotypic consequences of PYCRs depletion could not be rescued by external supplementation with either proline or nucleotides. Overall, our data suggest that, despite the mechanisms underlying the role of proline metabolism in colorectal tumorigenesis remain elusive, targeting the proline biosynthesis pathway is a suitable approach for the development of novel anti-cancer therapies.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
Screening a knowledge-based library of low molecular weight compounds against the proline biosynthetic enzyme 1-pyrroline-5-carboxylate 1 (PYCR1).Protein Sci. 2024 Jul;33(7):e5072. doi: 10.1002/pro.5072. Protein Sci. 2024. PMID: 39133178
-
Deciphering the Effects of the PYCR Family on Cell Function, Prognostic Value, Immune Infiltration in ccRCC and Pan-Cancer.Int J Mol Sci. 2024 Jul 25;25(15):8096. doi: 10.3390/ijms25158096. Int J Mol Sci. 2024. PMID: 39125668 Free PMC article.
-
Human mitochondrial pyrroline-5-carboxylate reductase 1 promotes invasiveness and impacts survival in breast cancers.Carcinogenesis. 2017 May 1;38(5):519-531. doi: 10.1093/carcin/bgx022. Carcinogenesis. 2017. PMID: 28379297
-
Structure, biochemistry, and gene expression patterns of the proline biosynthetic enzyme pyrroline-5-carboxylate reductase (PYCR), an emerging cancer therapy target.Amino Acids. 2021 Dec;53(12):1817-1834. doi: 10.1007/s00726-021-02999-5. Epub 2021 May 18. Amino Acids. 2021. PMID: 34003320 Free PMC article. Review.
-
PYCR, a key enzyme in proline metabolism, functions in tumorigenesis.Amino Acids. 2021 Dec;53(12):1841-1850. doi: 10.1007/s00726-021-03047-y. Epub 2021 Jul 17. Amino Acids. 2021. PMID: 34273023 Review.
Cited by
-
Functional Impact of a Cancer-Related Variant in Human Δ1-Pyrroline-5-Carboxylate Reductase 1.ACS Omega. 2023 Jan 10;8(3):3509-3519. doi: 10.1021/acsomega.2c07788. eCollection 2023 Jan 24. ACS Omega. 2023. PMID: 36713721 Free PMC article.
-
Screening a knowledge-based library of low molecular weight compounds against the proline biosynthetic enzyme 1-pyrroline-5-carboxylate 1 (PYCR1).Protein Sci. 2024 Jul;33(7):e5072. doi: 10.1002/pro.5072. Protein Sci. 2024. PMID: 39133178
-
Roles of circRNA dysregulation in esophageal squamous cell carcinoma tumor microenvironment.Front Oncol. 2023 Jun 13;13:1153207. doi: 10.3389/fonc.2023.1153207. eCollection 2023. Front Oncol. 2023. PMID: 37384299 Free PMC article. Review.
-
Circ_0000705 facilitates proline metabolism of esophageal squamous cell carcinoma cells by targeting miR-621/PYCR1 axis.Discov Oncol. 2022 Jun 22;13(1):50. doi: 10.1007/s12672-022-00513-1. Discov Oncol. 2022. PMID: 35731336 Free PMC article.
-
Loss of mitochondrial pyruvate carrier 1 supports proline-dependent proliferation and collagen biosynthesis in ovarian cancer.Mol Metab. 2024 Mar;81:101900. doi: 10.1016/j.molmet.2024.101900. Epub 2024 Feb 13. Mol Metab. 2024. PMID: 38354856 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

