Molecular Tension Probes to Quantify Cell-Generated Mechanical Forces
- PMID: 35114645
- PMCID: PMC8819489
- DOI: 10.14348/molcells.2022.2049
Molecular Tension Probes to Quantify Cell-Generated Mechanical Forces
Abstract
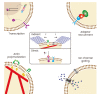
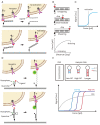
Living cells generate, sense, and respond to mechanical forces through their interaction with neighboring cells or extracellular matrix, thereby regulating diverse cellular processes such as growth, motility, differentiation, and immune responses. Dysregulation of mechanosensitive signaling pathways is found associated with the development and progression of various diseases such as cancer. Yet, little is known about the mechanisms behind mechano-regulation, largely due to the limited availability of tools to study it at the molecular level. The recent development of molecular tension probes allows measurement of cellular forces exerted by single ligandreceptor interaction, which has helped in revealing the hitherto unknown mechanistic details of various mechanosensitive processes in living cells. Here, we provide an introductory overview of two methods based on molecular tension probes, tension gauge tether (TGT), and molecular tension fluorescence microscopy (MTFM). TGT utilizes the irreversible rupture of double-stranded DNA tether upon application of force in the piconewton (pN) range, whereas MTFM utilizes the reversible extension of molecular springs such as polymer or single-stranded DNA hairpin under applied pN forces. Specifically, the underlying principle of how molecular tension probes measure cell-generated mechanical forces and their applications to mechanosensitive biological processes are described.
Keywords: cellular forces; mechanobiology; molecular spring; molecular tension fluorescence microscopy; tension gauge tether; tension probes.
Conflict of interest statement
The authors have no potential conflicts of interest to disclose.
Figures
Similar articles
-
Molecular Tension Probes for Imaging Forces at the Cell Surface.Acc Chem Res. 2017 Dec 19;50(12):2915-2924. doi: 10.1021/acs.accounts.7b00305. Epub 2017 Nov 21. Acc Chem Res. 2017. PMID: 29160067 Free PMC article. Review.
-
All-Covalent Nuclease-Resistant and Hydrogel-Tethered DNA Hairpin Probes Map pN Cell Traction Forces.ACS Appl Mater Interfaces. 2023 Jul 19;15(28):33362-33372. doi: 10.1021/acsami.3c04826. Epub 2023 Jul 6. ACS Appl Mater Interfaces. 2023. PMID: 37409737 Free PMC article.
-
Tension Gauge Tether Probes for Quantifying Growth Factor Mediated Integrin Mechanics and Adhesion.J Vis Exp. 2022 Feb 11;(180). doi: 10.3791/63529. J Vis Exp. 2022. PMID: 35225264
-
DNA Tension Probes to Map the Transient Piconewton Receptor Forces by Immune Cells.J Vis Exp. 2021 Mar 20;(169):10.3791/62348. doi: 10.3791/62348. J Vis Exp. 2021. PMID: 33818569 Free PMC article.
-
Investigating piconewton forces in cells by FRET-based molecular force microscopy.J Struct Biol. 2017 Jan;197(1):37-42. doi: 10.1016/j.jsb.2016.03.011. Epub 2016 Mar 12. J Struct Biol. 2017. PMID: 26980477 Review.
Cited by
-
Emerging Biophysics Tools for Biologists.Mol Cells. 2022 Jan 31;45(1):4-5. doi: 10.14348/molcells.2022.5001. Mol Cells. 2022. PMID: 35114643 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

