Artificial microRNA guide strand selection from duplexes with no mismatches shows a purine-rich preference for virus- and non-virus-based expression vectors in plants
- PMID: 35113475
- PMCID: PMC9129084
- DOI: 10.1111/pbi.13786
Artificial microRNA guide strand selection from duplexes with no mismatches shows a purine-rich preference for virus- and non-virus-based expression vectors in plants
Abstract
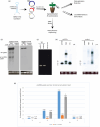
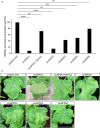
Artificial microRNA (amiRNA) technology has allowed researchers to direct efficient silencing of specific transcripts using as few as 21 nucleotides (nt). However, not all the artificially designed amiRNA constructs result in selection of the intended ~21-nt guide strand amiRNA. Selection of the miRNA guide strand from the mature miRNA duplex has been studied in detail in human and insect systems, but not so much for plants. Here, we compared a nuclear-replicating DNA viral vector (tomato mottle virus, ToMoV, based), a cytoplasmic-replicating RNA viral vector (tobacco mosaic virus, TMV, based), and a non-viral binary vector to express amiRNAs in plants. We then used deep sequencing and mutational analysis and show that when the structural factors caused by base mismatches in the mature amiRNA duplex were excluded, the nucleotide composition of the mature amiRNA region determined the guide strand selection. We found that the strand with excess purines was preferentially selected as the guide strand and the artificial miRNAs that had no mismatches in the amiRNA duplex were predominantly loaded into AGO2 instead of loading into AGO1 like the majority of the plant endogenous miRNAs. By performing assays for target effects, we also showed that only when the intended strand was selected as the guide strand and showed AGO loading, the amiRNA could provide the expected RNAi effects. Thus, by removing mismatches in the mature amiRNA duplex and designing the intended guide strand to contain excess purines provide better control of the guide strand selection of amiRNAs for functional RNAi effects.
Keywords: artificial microRNA; guide strand selection; small RNA deep sequencing; virus-based expression vector.
© 2022 The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
The Arabidopsis thaliana double-stranded RNA binding protein DRB1 directs guide strand selection from microRNA duplexes.RNA. 2009 Dec;15(12):2219-35. doi: 10.1261/rna.1646909. Epub 2009 Oct 27. RNA. 2009. PMID: 19861421 Free PMC article.
-
Artificial microRNA-mediated resistance against the monopartite begomovirus Cotton leaf curl Burewala virus.Virol J. 2013 Jul 12;10:231. doi: 10.1186/1743-422X-10-231. Virol J. 2013. PMID: 23844988 Free PMC article.
-
Evaluation and identification of candidate genes for artificial microRNA-mediated resistance to tomato spotted wilt virus.Virus Res. 2016 Jan 4;211:151-8. doi: 10.1016/j.virusres.2015.10.003. Epub 2015 Nov 11. Virus Res. 2016. PMID: 26454192
-
Artificial miRNAs as therapeutic tools: Challenges and opportunities.Wiley Interdiscip Rev RNA. 2021 Jul;12(4):e1640. doi: 10.1002/wrna.1640. Epub 2021 Jan 1. Wiley Interdiscip Rev RNA. 2021. PMID: 33386705 Review.
-
Artificial microRNA mediated gene silencing in plants: progress and perspectives.Plant Mol Biol. 2014 Sep;86(1-2):1-18. doi: 10.1007/s11103-014-0224-7. Epub 2014 Jul 15. Plant Mol Biol. 2014. PMID: 25022825 Review.
Cited by
-
The Intersection of Non-Coding RNAs Contributes to Forest Trees' Response to Abiotic Stress.Int J Mol Sci. 2022 Jun 7;23(12):6365. doi: 10.3390/ijms23126365. Int J Mol Sci. 2022. PMID: 35742808 Free PMC article. Review.
-
Transgene-free, virus-based gene silencing in plants by artificial microRNAs derived from minimal precursors.Nucleic Acids Res. 2023 Oct 27;51(19):10719-10736. doi: 10.1093/nar/gkad747. Nucleic Acids Res. 2023. PMID: 37713607 Free PMC article.
References
-
- Bartel, D.P . (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell, 116, 281–297. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

