Selective reduction of astrocyte apoE3 and apoE4 strongly reduces Aβ accumulation and plaque-related pathology in a mouse model of amyloidosis
- PMID: 35109920
- PMCID: PMC8811969
- DOI: 10.1186/s13024-022-00516-0
Selective reduction of astrocyte apoE3 and apoE4 strongly reduces Aβ accumulation and plaque-related pathology in a mouse model of amyloidosis
Abstract
Background: One of the key pathological hallmarks of Alzheimer disease (AD) is the accumulation of the amyloid-β (Aβ) peptide into amyloid plaques. The apolipoprotein E (APOE) gene is the strongest genetic risk factor for late-onset AD and has been shown to influence the accumulation of Aβ in the brain in an isoform-dependent manner. ApoE can be produced by different cell types in the brain, with astrocytes being the largest producer of apoE, although reactive microglia also express high levels of apoE. While studies have shown that altering apoE levels in the brain can influence the development of Aβ plaque pathology, it is not fully known how apoE produced by specific cell types, such as astrocytes, contributes to amyloid pathology.
Methods: We utilized APOE knock-in mice capable of having APOE selectively removed from astrocytes in a tamoxifen-inducible manner and crossed them with the APP/PS1-21 mouse model of amyloidosis. We analyzed the changes to Aβ plaque levels and assessed the impact on cellular responses to Aβ plaques when astrocytic APOE is removed.
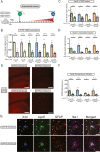
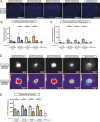
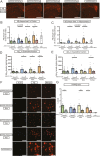
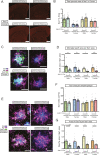
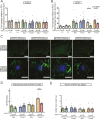
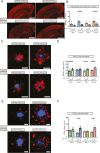
Results: Tamoxifen administration was capable of strongly reducing apoE levels in the brain by markedly reducing astrocyte apoE, while microglial apoE expression remained. Reduction of astrocytic apoE3 and apoE4 led to a large decrease in Aβ plaque deposition and less compact plaques. While overall Iba1+ microglia were unchanged in the cortex after reducing astrocyte apoE, the expression of the disease-associated microglial markers Clec7a and apoE were lower around amyloid plaques, indicating decreased microglial activation. Additionally, astrocyte GFAP levels are unchanged around amyloid plaques, but overall GFAP levels are reduced in the cortex of female apoE4 mice after a reduction in astrocytic apoE. Finally, while the amount of neuritic dystrophy around remaining individual plaques was increased with the removal of astrocytic apoE, the overall amount of cortical amyloid-associated neuritic dystrophy was significantly decreased.
Conclusion: This study reveals an important role of astrocytic apoE3 and apoE4 on the deposition and accumulation of Aβ plaques as well as on certain Aβ-associated downstream effects.
Keywords: Aldh1l1-Cre; Alzheimer disease; Amyloid; Apolipoprotein E; Astrocyte; Aβ; apoE.
© 2022. The Author(s).
Conflict of interest statement
D.M.H. is as an inventor on a patent licensed by Washington University to C2N Diagnostics on the therapeutic use of anti-tau antibodies. D.M.H. co-founded and is on the scientific advisory board of C2N Diagnostics. C2N Diagnostics has licensed certain anti-tau antibodies to AbbVie for therapeutic development. D.M.H. is on the scientific advisory board of Denali and consults for Genentech, Merck, and Cajal Neuroscience. All other authors have no competing interests.
Figures
Similar articles
-
Human APOE4 increases microglia reactivity at Aβ plaques in a mouse model of Aβ deposition.J Neuroinflammation. 2014 Jun 19;11:111. doi: 10.1186/1742-2094-11-111. J Neuroinflammation. 2014. PMID: 24948358 Free PMC article.
-
Trem2 deficiency differentially affects phenotype and transcriptome of human APOE3 and APOE4 mice.Mol Neurodegener. 2020 Jul 23;15(1):41. doi: 10.1186/s13024-020-00394-4. Mol Neurodegener. 2020. PMID: 32703241 Free PMC article.
-
Astrocytic APOE4 removal confers cerebrovascular protection despite increased cerebral amyloid angiopathy.Mol Neurodegener. 2023 Mar 16;18(1):17. doi: 10.1186/s13024-023-00610-x. Mol Neurodegener. 2023. PMID: 36922879 Free PMC article.
-
The interaction of amyloid-beta with ApoE.Subcell Biochem. 2005;38:255-72. doi: 10.1007/0-387-23226-5_13. Subcell Biochem. 2005. PMID: 15709483 Review.
-
The nucleation growth and reversibility of Amyloid-beta deposition in vivo.J Alzheimers Dis. 2006 Nov;10(2-3):291-301. doi: 10.3233/jad-2006-102-313. J Alzheimers Dis. 2006. PMID: 17119294 Review.
Cited by
-
Gut microbiota-astrocyte axis: new insights into age-related cognitive decline.Neural Regen Res. 2025 Apr 1;20(4):990-1008. doi: 10.4103/NRR.NRR-D-23-01776. Epub 2024 Apr 16. Neural Regen Res. 2025. PMID: 38989933 Free PMC article.
-
The association between dietary inflammatory index and cognitive function in adults with/without chronic kidney disease.Front Nutr. 2023 Nov 21;10:1279721. doi: 10.3389/fnut.2023.1279721. eCollection 2023. Front Nutr. 2023. PMID: 38075216 Free PMC article.
-
The gut microbiome in Alzheimer's disease: what we know and what remains to be explored.Mol Neurodegener. 2023 Feb 1;18(1):9. doi: 10.1186/s13024-023-00595-7. Mol Neurodegener. 2023. PMID: 36721148 Free PMC article. Review.
-
Anti-inflammatory clearance of amyloid-β by a chimeric Gas6 fusion protein.Nat Med. 2022 Sep;28(9):1802-1812. doi: 10.1038/s41591-022-01926-9. Epub 2022 Aug 4. Nat Med. 2022. PMID: 35927581
-
Antibody Assay and Anti-Inflammatory Function Evaluation of Therapeutic Potential of Different Intravenous Immunoglobulins for Alzheimer's Disease.Int J Mol Sci. 2023 Mar 14;24(6):5549. doi: 10.3390/ijms24065549. Int J Mol Sci. 2023. PMID: 36982622 Free PMC article.
References
-
- 2021 Alzheimer’s disease facts and figures. Alzheimers Dement. 2021;17:327–406. https://pubmed.ncbi.nlm.nih.gov/33756057/. - PubMed
-
- International AD. Guerchet M, Prince M. Numbers of people with dementia around the world. 2020.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

