Anastasis Drives Senescence and Non-Cell Autonomous Neurodegeneration in the Astrogliopathy Alexander Disease
- PMID: 35105675
- PMCID: PMC8944235
- DOI: 10.1523/JNEUROSCI.1659-21.2021
Anastasis Drives Senescence and Non-Cell Autonomous Neurodegeneration in the Astrogliopathy Alexander Disease
Abstract
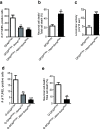
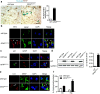
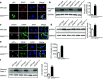
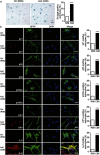
Anastasis is a recently described process in which cells recover after late-stage apoptosis activation. The functional consequences of anastasis for cells and tissues are not clearly understood. Using Drosophila, rat and human cells and tissues, including analyses of both males and females, we present evidence that glia undergoing anastasis in the primary astrogliopathy Alexander disease subsequently express hallmarks of senescence. These senescent glia promote non-cell autonomous death of neurons by secreting interleukin family cytokines. Our findings demonstrate that anastasis can be dysfunctional in neurologic disease by inducing a toxic senescent population of astroglia.SIGNIFICANCE STATEMENT Under some conditions cells otherwise destined to die can be rescued just before death in a process called anastasis, or "rising from the dead." The fate and function of cells undergoing a near death experience is not well understood. Here, we find that in models and patient cells from Alexander disease, an important brain disorder in which glial cells promote neuronal dysfunction and death, anastasis of astrocytic glia leads to secretion of toxic signaling molecules and neurodegeneration. These studies demonstrate a previously unexpected deleterious consequence of rescuing cells on the brink of death and suggest therapeutic strategies for Alexander disease and related disorders of glia.
Copyright © 2022 the authors.
Figures

Similar articles
-
Detecting Anastasis In Vivo by CaspaseTracker Biosensor.J Vis Exp. 2018 Feb 1;(132):54107. doi: 10.3791/54107. J Vis Exp. 2018. PMID: 29443051 Free PMC article.
-
Protein misfolding and oxidative stress promote glial-mediated neurodegeneration in an Alexander disease model.J Neurosci. 2011 Feb 23;31(8):2868-77. doi: 10.1523/JNEUROSCI.3410-10.2011. J Neurosci. 2011. PMID: 21414908 Free PMC article.
-
Anastasis: cell recovery mechanisms and potential role in cancer.Cell Commun Signal. 2022 Jun 3;20(1):81. doi: 10.1186/s12964-022-00880-w. Cell Commun Signal. 2022. PMID: 35659306 Free PMC article. Review.
-
Nitric oxide mediates glial-induced neurodegeneration in Alexander disease.Nat Commun. 2015 Nov 26;6:8966. doi: 10.1038/ncomms9966. Nat Commun. 2015. PMID: 26608817 Free PMC article.
-
Anastasis: recovery from the brink of cell death.R Soc Open Sci. 2018 Sep 19;5(9):180442. doi: 10.1098/rsos.180442. eCollection 2018 Sep. R Soc Open Sci. 2018. PMID: 30839720 Free PMC article. Review.
Cited by
-
Large-scale gene expression changes in APP/PSEN1 and GFAP mutation models exhibit high congruence with Alzheimer's disease.PLoS One. 2024 Jan 18;19(1):e0291995. doi: 10.1371/journal.pone.0291995. eCollection 2024. PLoS One. 2024. PMID: 38236817 Free PMC article.
-
Cell survival following direct executioner-caspase activation.Proc Natl Acad Sci U S A. 2023 Jan 24;120(4):e2216531120. doi: 10.1073/pnas.2216531120. Epub 2023 Jan 20. Proc Natl Acad Sci U S A. 2023. PMID: 36669100 Free PMC article.
References
-
- Brenner M, Goldman JE, Quinlan RA, Messing A (2009) Alexander disease: a genetic disorder of astrocytes. In: Astrocytes in (patho)physiology of the nervous system, pp 591–648. New York: Springer.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
