Duplex DNA and BLM regulate gate opening by the human TopoIIIα-RMI1-RMI2 complex
- PMID: 35102151
- PMCID: PMC8803869
- DOI: 10.1038/s41467-022-28082-5
Duplex DNA and BLM regulate gate opening by the human TopoIIIα-RMI1-RMI2 complex
Abstract
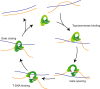
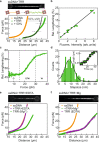
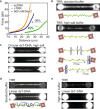
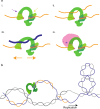
Topoisomerase IIIα is a type 1A topoisomerase that forms a complex with RMI1 and RMI2 called TRR in human cells. TRR plays an essential role in resolving DNA replication and recombination intermediates, often alongside the helicase BLM. While the TRR catalytic cycle is known to involve a protein-mediated single-stranded (ss)DNA gate, the detailed mechanism is not fully understood. Here, we probe the catalytic steps of TRR using optical tweezers and fluorescence microscopy. We demonstrate that TRR forms an open gate in ssDNA of 8.5 ± 3.8 nm, and directly visualize binding of a second ssDNA or double-stranded (ds)DNA molecule to the open TRR-ssDNA gate, followed by catenation in each case. Strikingly, dsDNA binding increases the gate size (by ~16%), while BLM alters the mechanical flexibility of the gate. These findings reveal an unexpected plasticity of the TRR-ssDNA gate size and suggest that TRR-mediated transfer of dsDNA may be more relevant in vivo than previously believed.
© 2022. The Author(s).
Conflict of interest statement
The combined optical tweezers and fluorescence technologies used in this article are patented and licensed to LUMICKS B.V., in which E.J.G.P. and G.J.L.W. have a financial interest. The remaining authors declare no competing interests.
Figures



Similar articles
-
BLM and RMI1 alleviate RPA inhibition of TopoIIIα decatenase activity.PLoS One. 2012;7(7):e41208. doi: 10.1371/journal.pone.0041208. Epub 2012 Jul 20. PLoS One. 2012. PMID: 22911760 Free PMC article.
-
Multifaceted role of the Topo IIIα-RMI1-RMI2 complex and DNA2 in the BLM-dependent pathway of DNA break end resection.Nucleic Acids Res. 2014;42(17):11083-91. doi: 10.1093/nar/gku803. Epub 2014 Sep 8. Nucleic Acids Res. 2014. PMID: 25200081 Free PMC article.
-
Monopolar spindle 1 (MPS1) protein-dependent phosphorylation of RecQ-mediated genome instability protein 2 (RMI2) at serine 112 is essential for BLM-Topo III α-RMI1-RMI2 (BTR) protein complex function upon spindle assembly checkpoint (SAC) activation during mitosis.J Biol Chem. 2013 Nov 22;288(47):33500-33508. doi: 10.1074/jbc.M113.470823. Epub 2013 Oct 9. J Biol Chem. 2013. PMID: 24108125 Free PMC article.
-
Unravelling the mechanisms of Type 1A topoisomerases using single-molecule approaches.Nucleic Acids Res. 2021 Jun 4;49(10):5470-5492. doi: 10.1093/nar/gkab239. Nucleic Acids Res. 2021. PMID: 33963870 Free PMC article. Review.
-
The dissolution of double Holliday junctions.Cold Spring Harb Perspect Biol. 2014 Jul 1;6(7):a016477. doi: 10.1101/cshperspect.a016477. Cold Spring Harb Perspect Biol. 2014. PMID: 24984776 Free PMC article. Review.
Cited by
-
TOP3A coupling with replication forks and repair of TOP3A cleavage complexes.Cell Cycle. 2024 Jan;23(2):115-130. doi: 10.1080/15384101.2024.2314440. Epub 2024 Feb 11. Cell Cycle. 2024. PMID: 38341866 Free PMC article. Review.
-
PARP1-driven repair of topoisomerase IIIα DNA-protein crosslinks by FEN1.Cell Rep. 2024 Aug 27;43(8):114522. doi: 10.1016/j.celrep.2024.114522. Epub 2024 Jul 18. Cell Rep. 2024. PMID: 39028621 Free PMC article.
-
Replication-associated formation and repair of human topoisomerase IIIα cleavage complexes.Nat Commun. 2023 Apr 6;14(1):1925. doi: 10.1038/s41467-023-37498-6. Nat Commun. 2023. PMID: 37024461 Free PMC article.
-
What's on the Other Side of the Gate: A Structural Perspective on DNA Gate Opening of Type IA and IIA DNA Topoisomerases.Int J Mol Sci. 2023 Feb 16;24(4):3986. doi: 10.3390/ijms24043986. Int J Mol Sci. 2023. PMID: 36835394 Free PMC article. Review.
-
The interaction between transport-segment DNA and topoisomerase IA-crystal structure of MtbTOP1 in complex with both G- and T-segments.Nucleic Acids Res. 2023 Jan 11;51(1):349-364. doi: 10.1093/nar/gkac1205. Nucleic Acids Res. 2023. PMID: 36583363 Free PMC article.
References
-
- Champoux JJ. DNA Topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 2001;70:369–413. - PubMed
-
- Li Z, Mondragón A, Hiasa H, Marians KJ, DiGate RJ. Identification of a unique domain essential for Escherichia coli DNA topoisomerase III-catalysed decatenation of replication intermediates. Mol. Microbiol. 2000;35:888–895. - PubMed
-
- Mondragón A, DiGate R. The structure of Escherichia coli DNA topoisomerase III. Structure. 1999;7:1373–1383. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

