Response of Multiple Tissues to Drought Revealed by a Weighted Gene Co-Expression Network Analysis in Foxtail Millet [ Setaria italica (L.) P. Beauv.]
- PMID: 35095942
- PMCID: PMC8790073
- DOI: 10.3389/fpls.2021.746166
Response of Multiple Tissues to Drought Revealed by a Weighted Gene Co-Expression Network Analysis in Foxtail Millet [ Setaria italica (L.) P. Beauv.]
Abstract
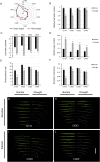
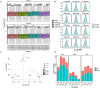
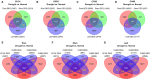
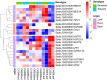
Characterization of drought-tolerance mechanisms during the jointing stage in foxtail millet under water-limited conditions is essential for improving the grain yield of this C4 crop species. In this trial, two drought-tolerant and two drought-sensitive cultivars were examined using transcriptomic dissections of three tissues (root, stem, and leaf) under naturally occurring water-limited conditions. We detected a total of 32,170 expressed genes and characterized 13,552 differentially expressed genes (DEGs) correlated with drought treatment. The majority of DEGs were identified in the root tissue, followed by leaf and stem tissues, and the number of DEGs identified in the stems of drought-sensitive cultivars was about two times higher than the drought-tolerant ones. A total of 127 differentially expressed transcription factors (DETFs) with different drought-responsive patterns were identified between drought-tolerant and drought-sensitive genotypes (including MYB, b-ZIP, ERF, and WRKY). Furthermore, a total of 34 modules were constructed for all expressed genes using a weighted gene co-expression network analysis (WGCNA), and seven modules were closely related to the drought treatment. A total of 1,343 hub genes (including RAB18, LEA14, and RD22) were detected in the drought-related module, and cell cycle and DNA replication-related transcriptional pathways were identified as vital regulators of drought tolerance in foxtail millet. The results of this study provide a comprehensive overview of how Setaria italica copes with drought-inflicted environments during the jointing stage through transcriptional regulating strategies in different organs and lays a foundation for the improvement of drought-tolerant cereal cultivars through genomic editing approaches in the future.
Keywords: RNA-Seq; Setaria italica; WGCNA; drought; jointing stage; multi-tissue.
Copyright © 2022 Zhang, Zhi, Li, Guo, Feng, Tang, Guo, Zhang, Jia and Diao.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Comparative metabolomic and transcriptomic analysis reveals a coexpression network of the carotenoid metabolism pathway in the panicle of Setaria italica.BMC Plant Biol. 2022 Mar 8;22(1):105. doi: 10.1186/s12870-022-03467-2. BMC Plant Biol. 2022. PMID: 35260077 Free PMC article.
-
An investigation into the beneficial effects and molecular mechanisms of humic acid on foxtail millet under drought conditions.PLoS One. 2020 Jun 2;15(6):e0234029. doi: 10.1371/journal.pone.0234029. eCollection 2020. PLoS One. 2020. PMID: 32484836 Free PMC article.
-
Genome-Wide Gene Expression Profiles Analysis Reveal Novel Insights into Drought Stress in Foxtail Millet (Setaria italica L.).Int J Mol Sci. 2020 Nov 12;21(22):8520. doi: 10.3390/ijms21228520. Int J Mol Sci. 2020. PMID: 33198267 Free PMC article.
-
Transcriptome analysis and identification of the low potassium stress-responsive gene SiSnRK2.6 in foxtail millet (Setaria italica L.).Theor Appl Genet. 2024 Jan 16;137(1):22. doi: 10.1007/s00122-023-04532-6. Theor Appl Genet. 2024. PMID: 38227064 Review.
-
Adaptation of Foxtail Millet (Setaria italica L.) to Abiotic Stresses: A Special Perspective of Responses to Nitrogen and Phosphate Limitations.Front Plant Sci. 2020 Feb 28;11:187. doi: 10.3389/fpls.2020.00187. eCollection 2020. Front Plant Sci. 2020. PMID: 32184798 Free PMC article. Review.
Cited by
-
Comparative Analysis of Untargeted Metabolomics in Tolerant and Sensitive Genotypes of Common Bean (Phaseolus vulgaris L.) Seeds Exposed to Terminal Drought Stress.Metabolites. 2022 Oct 5;12(10):944. doi: 10.3390/metabo12100944. Metabolites. 2022. PMID: 36295847 Free PMC article.
-
Comparative Transcriptome Analysis of CMV or 2b-Deficient CMV-Infected dcl2dcl4 Reveals the Effects of Viral Infection on Symptom Induction in Arabidopsis thaliana.Viruses. 2022 Jul 21;14(7):1582. doi: 10.3390/v14071582. Viruses. 2022. PMID: 35891562 Free PMC article.
-
Identification and expression analysis of bZIP transcription factors in Setaria italica in response to dehydration stress.Front Genet. 2024 Aug 30;15:1466486. doi: 10.3389/fgene.2024.1466486. eCollection 2024. Front Genet. 2024. PMID: 39280094 Free PMC article.
-
Expression of Foxtail Millet bZIP Transcription Factor SibZIP67 Enhances Drought Tolerance in Arabidopsis.Biomolecules. 2024 Aug 7;14(8):958. doi: 10.3390/biom14080958. Biomolecules. 2024. PMID: 39199345 Free PMC article.
-
Drought Responses in Poaceae: Exploring the Core Components of the ABA Signaling Pathway in Setaria italica and Setaria viridis.Plants (Basel). 2024 May 23;13(11):1451. doi: 10.3390/plants13111451. Plants (Basel). 2024. PMID: 38891260 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Miscellaneous

