Mechanisms for Improving Hepatic Glucolipid Metabolism by Cinnamic Acid and Cinnamic Aldehyde: An Insight Provided by Multi-Omics
- PMID: 35087857
- PMCID: PMC8786797
- DOI: 10.3389/fnut.2021.794841
Mechanisms for Improving Hepatic Glucolipid Metabolism by Cinnamic Acid and Cinnamic Aldehyde: An Insight Provided by Multi-Omics
Abstract
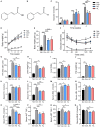
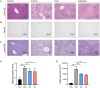
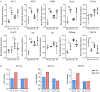
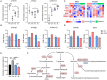
Cinnamic acid (AC) and cinnamic aldehyde (AL) are two chemicals enriched in cinnamon and have been previously proved to improve glucolipid metabolism, thus ameliorating metabolic disorders. In this study, we employed transcriptomes and proteomes on AC and AL treated db/db mice in order to explore the underlying mechanisms for their effects. Db/db mice were divided into three groups: the control group, AC group and AL group. Gender- and age-matched wt/wt mice were used as a normal group. After 4 weeks of treatments, mice were sacrificed, and liver tissues were used for further analyses. Functional enrichment of differentially expressed genes (DEGs) and differentially expressed proteins (DEPs) were performed using Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases. DEPs were further verified by parallel reaction monitoring (PRM). The results suggested that AC and AL share similar mechanisms, and they may improve glucolipid metabolism by improving mitochondrial functions, decreasing serotonin contents and upregulating autophagy mediated lipid clearance. This study provides an insight into the molecular mechanisms of AC and AL on hepatic transcriptomes and proteomes in disrupted metabolic situations and lays a foundation for future experiments.
Keywords: cinnamaldehyde; cinnamic acid; db/db; glucolipid metabolism; liver; proteome; transcriptome.
Copyright © 2022 Wu, Wang, Yang, Qin, Qin, Hu, Zhang, Sun, Ding, Wu and Liu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Hypoglycemic mechanisms of Ganoderma lucidum polysaccharides F31 in db/db mice via RNA-seq and iTRAQ.Food Funct. 2018 Dec 13;9(12):6495-6507. doi: 10.1039/c8fo01656a. Food Funct. 2018. PMID: 30467564
-
Comparative Proteomic Analysis of Liver Tissues and Serum in db/db Mice.Int J Mol Sci. 2022 Aug 26;23(17):9687. doi: 10.3390/ijms23179687. Int J Mol Sci. 2022. PMID: 36077090 Free PMC article.
-
Integration of transcriptomic and proteomic analyses for finger millet [Eleusine coracana (L.) Gaertn.] in response to drought stress.PLoS One. 2021 Feb 17;16(2):e0247181. doi: 10.1371/journal.pone.0247181. eCollection 2021. PLoS One. 2021. PMID: 33596255 Free PMC article.
-
Hepatic transcriptome and proteome analyses provide new insights into the regulator mechanism of dietary avicularin in diabetic mice.Food Res Int. 2019 Nov;125:108570. doi: 10.1016/j.foodres.2019.108570. Epub 2019 Jul 19. Food Res Int. 2019. PMID: 31554135
-
Cinnamaldehyde in diabetes: A review of pharmacology, pharmacokinetics and safety.Pharmacol Res. 2017 Aug;122:78-89. doi: 10.1016/j.phrs.2017.05.019. Epub 2017 May 27. Pharmacol Res. 2017. PMID: 28559210 Review.
Cited by
-
Regulation Mechanism and Potential Value of Active Substances in Spices in Alcohol-Liver-Intestine Axis Health.Int J Mol Sci. 2024 Mar 27;25(7):3728. doi: 10.3390/ijms25073728. Int J Mol Sci. 2024. PMID: 38612538 Free PMC article. Review.
-
Cinnamaldehyde supplementation acts as an insulin mimetic compound improving glucose metabolism during adolescence, but not during adulthood, in healthy male rats.Hormones (Athens). 2023 Jun;22(2):295-304. doi: 10.1007/s42000-023-00442-w. Epub 2023 Feb 22. Hormones (Athens). 2023. PMID: 36810755
References
-
- Saeedi P, Petersohn I, Salpea P, Malanda B, Karuranga S, Unwin N, et al. . Global and regional diabetes prevalence estimates for 2019 and projections for 2030 and 2045: Results from the International Diabetes Federation Diabetes Atlas, 9(th) edition. Diabetes Res Clin Pract. (2019) 157:107843. 10.1016/j.diabres.2019.107843 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

