A high-throughput assay for directly monitoring nucleolar rRNA biogenesis
- PMID: 35078352
- PMCID: PMC8790372
- DOI: 10.1098/rsob.210305
A high-throughput assay for directly monitoring nucleolar rRNA biogenesis
Abstract
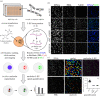
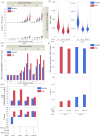
Studies of the regulation of nucleolar function are critical for ascertaining clearer insights into the basic biological underpinnings of ribosome biogenesis (RB), and for future development of therapeutics to treat cancer and ribosomopathies. A number of high-throughput primary assays based on morphological alterations of the nucleolus can indirectly identify hits affecting RB. However, there is a need for a more direct high-throughput assay for a nucleolar function to further evaluate hits. Previous reports have monitored nucleolar rRNA biogenesis using 5-ethynyl uridine (5-EU) in low-throughput. We report a miniaturized, high-throughput 5-EU assay that enables specific calculation of nucleolar rRNA biogenesis inhibition, based on co-staining of the nucleolar protein fibrillarin (FBL). The assay uses two siRNA controls: a negative non-targeting siRNA control and a positive siRNA control targeting RNA Polymerase 1 (RNAP1; POLR1A), and specifically quantifies median 5-EU signal within nucleoli. Maximum nuclear 5-EU signal can also be used to monitor the effects of putative small-molecule inhibitors of RNAP1, like BMH-21, or other treatment conditions that cause FBL dispersion. We validate the 5-EU assay on 68 predominately nucleolar hits from a high-throughput primary screen, showing that 58/68 hits significantly inhibit nucleolar rRNA biogenesis. Our new method establishes direct quantification of nucleolar function in high-throughput, facilitating closer study of RB in health and disease.
Keywords: RNA polymerase 1; high-throughput screening; nucleolus; pre-rRNA processing; pre-rRNA transcription; ribosome biogenesis.
Conflict of interest statement
We declare we have no competing interests.
Figures

Similar articles
-
rRNA transcription is integral to phase separation and maintenance of nucleolar structure.PLoS Genet. 2023 Aug 28;19(8):e1010854. doi: 10.1371/journal.pgen.1010854. eCollection 2023 Aug. PLoS Genet. 2023. PMID: 37639467 Free PMC article.
-
Nucleolar stress: From development to cancer.Semin Cell Dev Biol. 2023 Feb 28;136:64-74. doi: 10.1016/j.semcdb.2022.04.001. Epub 2022 Apr 8. Semin Cell Dev Biol. 2023. PMID: 35410715 Free PMC article. Review.
-
Diverse Regulators of Human Ribosome Biogenesis Discovered by Changes in Nucleolar Number.Cell Rep. 2018 Feb 13;22(7):1923-1934. doi: 10.1016/j.celrep.2018.01.056. Cell Rep. 2018. PMID: 29444442 Free PMC article.
-
The roles of RRP15 in nucleolar formation, ribosome biogenesis and checkpoint control in human cells.Oncotarget. 2017 Feb 21;8(8):13240-13252. doi: 10.18632/oncotarget.14658. Oncotarget. 2017. PMID: 28099941 Free PMC article.
-
Ribosomal RNA and nucleolar proteins from the oocyte are to some degree used for embryonic nucleolar formation in cattle and pig.Theriogenology. 2007 Sep 1;68 Suppl 1:S63-70. doi: 10.1016/j.theriogenology.2007.03.015. Epub 2007 Apr 26. Theriogenology. 2007. PMID: 17466364 Review.
Cited by
-
Discovery of novel microRNA mimic repressors of ribosome biogenesis.Nucleic Acids Res. 2024 Feb 28;52(4):1988-2011. doi: 10.1093/nar/gkad1235. Nucleic Acids Res. 2024. PMID: 38197221 Free PMC article.
-
Targeting Ribosome Biogenesis in Cancer: Lessons Learned and Way Forward.Cancers (Basel). 2022 Apr 24;14(9):2126. doi: 10.3390/cancers14092126. Cancers (Basel). 2022. PMID: 35565259 Free PMC article. Review.
-
Mapping subcellular localizations of unannotated microproteins and alternative proteins with MicroID.Mol Cell. 2022 Aug 4;82(15):2900-2911.e7. doi: 10.1016/j.molcel.2022.06.035. Epub 2022 Jul 28. Mol Cell. 2022. PMID: 35905735 Free PMC article.
-
The cytidine deaminase APOBEC3A regulates nucleolar function to promote cell growth and ribosome biogenesis.PLoS Biol. 2024 Jul 8;22(7):e3002718. doi: 10.1371/journal.pbio.3002718. eCollection 2024 Jul. PLoS Biol. 2024. PMID: 38976757 Free PMC article.
-
All these screens that we've done: how functional genetic screens have informed our understanding of ribosome biogenesis.Biosci Rep. 2023 Jul 26;43(7):BSR20230631. doi: 10.1042/BSR20230631. Biosci Rep. 2023. PMID: 37335083 Free PMC article. Review.
References
-
- Lafontaine DLJ, Riback JA, Bascetin R, Brangwynne CP. 2020. The nucleolus as a multiphase liquid condensate. Nat. Rev. Mol. Cell Biol. 22, 165-182. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

