The Roles of Floral Organ Genes in Regulating Rosaceae Fruit Development
- PMID: 35069608
- PMCID: PMC8766977
- DOI: 10.3389/fpls.2021.644424
The Roles of Floral Organ Genes in Regulating Rosaceae Fruit Development
Abstract
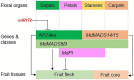
The function of floral organ identity genes, APETALA1/2/3, PISTILLATA, AGAMOUS, and SEPALLATA1/2/3, in flower development is highly conserved across angiosperms. Emerging evidence shows that these genes also play important roles in the development of the fruit that originates from floral organs following pollination and fertilization. However, their roles in fruit development may vary significantly between species depending on the floral organ types contributing to the fruit tissues. Fruits of the Rosaceae family develop from different floral organ types depending on the species, for example, peach fruit flesh develops from carpellary tissues, whereas apple and strawberry fruit flesh develop from extra-carpellary tissues, the hypanthium and receptacle, respectively. In this review, we summarize recent advances in understanding floral organ gene function in Rosaceae fruit development and analyze the similarities and diversities within this family as well as between Rosaceae and the model plant species Arabidopsis and tomato. We conclude by suggesting future research opportunities using genomics resources to rapidly dissect gene function in this family of perennial plants.
Keywords: AP2; MADS-box; apple; fruit development; miR172; peach; strawberry.
Copyright © 2022 Yao, Kang, Gu and Gleave.
Conflict of interest statement
Authors AG and J-LY were employed by company The New Zealand Institute for Plant and Food Research Limited. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Developmental Mechanisms of Fleshy Fruit Diversity in Rosaceae.Annu Rev Plant Biol. 2020 Apr 29;71:547-573. doi: 10.1146/annurev-arplant-111119-021700. Annu Rev Plant Biol. 2020. PMID: 32442388
-
Identification of genes preferentially expressed in wild strawberry receptacle fruit and demonstration of their promoter activities.Hortic Res. 2019 May 1;6:50. doi: 10.1038/s41438-019-0134-6. eCollection 2019. Hortic Res. 2019. PMID: 31044078 Free PMC article.
-
Comparative transcriptomic analysis of apple and peach fruits: insights into fruit type specification.Plant J. 2022 Mar;109(6):1614-1629. doi: 10.1111/tpj.15633. Epub 2021 Dec 27. Plant J. 2022. PMID: 34905278
-
The origin and evolution of carpels and fruits from an evo-devo perspective.J Integr Plant Biol. 2023 Feb;65(2):283-298. doi: 10.1111/jipb.13351. Epub 2023 Jan 2. J Integr Plant Biol. 2023. PMID: 36031801 Review.
-
Genomics of pear and other Rosaceae fruit trees.Breed Sci. 2016 Jan;66(1):148-59. doi: 10.1270/jsbbs.66.148. Epub 2016 Jan 1. Breed Sci. 2016. PMID: 27069399 Free PMC article. Review.
Cited by
-
The haplotype-resolved autotetraploid genome assembly provides insights into the genomic evolution and fruit divergence in wax apple (Syzygium samarangense (Blume) Merr. and Perry).Hortic Res. 2023 Oct 25;10(12):uhad214. doi: 10.1093/hr/uhad214. eCollection 2023 Dec. Hortic Res. 2023. PMID: 38077494 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources
Research Materials

