Particle-based analysis elucidates the real retention capacities of virus filters and enables optimal virus clearance study design with evaluation systems of diverse virological characteristics
- PMID: 35064964
- PMCID: PMC9285584
- DOI: 10.1002/btpr.3237
Particle-based analysis elucidates the real retention capacities of virus filters and enables optimal virus clearance study design with evaluation systems of diverse virological characteristics
Abstract
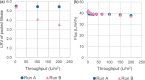
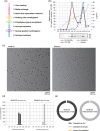
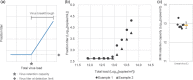
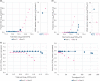
In virus clearance study (VCS) design, the amount of virus loaded onto the virus filters (VF) must be carefully controlled. A large amount of virus is required to demonstrate sufficient virus removal capability; however, too high a viral load causes virus breakthrough and reduces log reduction values. We have seen marked variation in the virus removal performance for VFs even with identical VCS design. Understanding how identical virus infectivity, materials and operating conditions can yield such different results is key to optimizing VCS design. The present study developed a particle number-based method for VCS and investigated the effects on VF performance of discrepancies between apparent virus amount and total particle number of minute virus of mice. Co-spiking of empty and genome-containing particles resulted in a decrease in the virus removal performance proportional to the co-spike ratio. This suggests that empty particles are captured in the same way as genome-containing particles, competing for retention capacity. In addition, between virus titration methods with about 2.0 Log10 difference in particle-to-infectivity ratios, there was a 20-fold decrease in virus retention capacity limiting the throughput that maintains the required LRV (e.g., 4.0), calculated using infectivity titers. These findings suggest that ignoring virus particle number in VCS design can cause virus overloading and accelerate filter breakthrough. This article asserts the importance of focusing on virus particle number and discusses optimization of VCS design that is unaffected by virological characteristics of evaluation systems and adequately reflect the VF retention capacity.
Keywords: minute virus of mice; particle to infectivity ratio; virus filter; virus filtration; virus retention capacity.
© 2022 Asahi Kasei Medical Co. Ltd. Biotechnology Progress published by Wiley Periodicals LLC on behalf of American Institute of Chemical Engineers.
Figures




Similar articles
-
Limits in virus filtration capability? Impact of virus quality and spike level on virus removal with xenotropic murine leukemia virus.Biotechnol Prog. 2015 Jan-Feb;31(1):135-44. doi: 10.1002/btpr.2020. Epub 2014 Nov 29. Biotechnol Prog. 2015. PMID: 25395156
-
Reproduction of influenza viruses; quantitative investigations with particle enumeration procedures on the dynamics of influenza A and B virus reproduction.J Exp Med. 1955 Oct 1;102(4):441-73. doi: 10.1084/jem.102.4.441. J Exp Med. 1955. PMID: 13263486 Free PMC article.
-
Development of a transient inline spiking system for evaluating virus clearance in continuous bioprocessing-Proof of concept for virus filtration.Biotechnol Bioeng. 2022 Aug;119(8):2134-2141. doi: 10.1002/bit.28119. Epub 2022 May 5. Biotechnol Bioeng. 2022. PMID: 35470427
-
Use of MMV as a Single Worst-Case Model Virus in Viral Filter Validation Studies.PDA J Pharm Sci Technol. 2014 May-Jun;68(3):297-311. doi: 10.5731/pdajpst.2014.00978. PDA J Pharm Sci Technol. 2014. PMID: 25188350 Review.
-
Upflow anaerobic sludge blanket reactor--a review.Indian J Environ Health. 2001 Apr;43(2):1-82. Indian J Environ Health. 2001. PMID: 12397675 Review.
References
-
- ICH Harmonized Tripartite Guideline Q5A (R1) . Viral safety evaluation of biotechnology products derived from cell lines of human or animal origin. US Fed Reg. 1999;63(185):51074. - PubMed
-
- Guideline on virus safety evaluation of biotechnological investigational medicinal products. EMA 2008: EMEA/CHMP/BWP/398498/2005;1–9. - PubMed
-
- Sterilizing filtration of liquids task force. sterilizing filtration of liquids. technical report no. 26 (revised 2008). PDA J Pharm Sci Technol. 2008;62(26):2‐60. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

