Novel Neutralizing Epitope of PEDV S1 Protein Identified by IgM Monoclonal Antibody
- PMID: 35062329
- PMCID: PMC8778753
- DOI: 10.3390/v14010125
Novel Neutralizing Epitope of PEDV S1 Protein Identified by IgM Monoclonal Antibody
Abstract
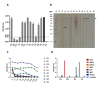
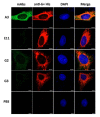
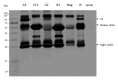
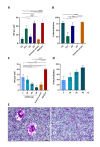
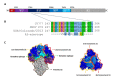
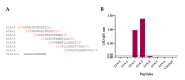
Porcine epidemic diarrhea virus (PEDV) causes devastating enteric disease that inflicts huge economic damage on the swine industry worldwide. A safe and highly effective PEDV vaccine that contains only the virus-neutralizing epitopes (not enhancing epitope), as well as a ready-to-use PEDV neutralizing antibody for the passive immunization of PEDV vulnerable piglets (during the first week of life) are needed, particularly for PEDV-endemic farms. In this study, we generated monoclonal antibodies (mAbs) to the recombinant S1 domain of PEDV spike (S) protein and tested their PEDV neutralizing activity by CPE-reduction assay. The mAb secreted by one hybrodoma clone (A3), that also bound to the native S1 counterpart from PEDV-infected cells (tested by combined co-immunoprecipitation and Western blotting), neutralized PEDV infectivity. Epitope of the neutralizing mAb (mAbA3) locates in the S1A subdomain of the spike protein, as identified by phage mimotope search and multiple sequence alignment, and peptide binding-ELISA. The newly identified epitope is shared by PEDV G1 and G2 strains and other alphacoronaviruses. In summary, mAbA3 may be useful as a ready-to-use antibody for passive immunization of PEDV-susceptible piglets, while the novel neutralizing epitope, together with other, previously known protective epitopes, have potential as an immunogenic cocktail for a safe, next-generation PEDV vaccine.
Keywords: monoclonal antibody; neutralization assay; neutralizing antibody; phage mimotope; porcine epidemic diarrhea virus (PEDV); spike (S) protein.
Conflict of interest statement
No potential conflict of interest was reported by the authors.
Figures


Similar articles
-
Cell Attachment Domains of the Porcine Epidemic Diarrhea Virus Spike Protein Are Key Targets of Neutralizing Antibodies.J Virol. 2017 May 26;91(12):e00273-17. doi: 10.1128/JVI.00273-17. Print 2017 Jun 15. J Virol. 2017. PMID: 28381581 Free PMC article.
-
Identification of a novel B-cell epitope in the spike protein of porcine epidemic diarrhea virus.Virol J. 2020 Apr 3;17(1):46. doi: 10.1186/s12985-020-01305-1. Virol J. 2020. PMID: 32245493 Free PMC article.
-
High-affinity monoclonal antibodies against the porcine epidemic diarrhea virus S1 protein.BMC Vet Res. 2024 Jun 3;20(1):239. doi: 10.1186/s12917-024-04091-y. BMC Vet Res. 2024. PMID: 38831363 Free PMC article.
-
Immuno-Colorimetric Neutralization Test: A Surrogate for Widely Used Plaque Reduction Neutralization Tests in Public Health Virology.Viruses. 2023 Apr 10;15(4):939. doi: 10.3390/v15040939. Viruses. 2023. PMID: 37112919 Free PMC article. Review.
-
Human Antibodies for Viral Infections.Annu Rev Immunol. 2022 Apr 26;40:349-386. doi: 10.1146/annurev-immunol-042718-041309. Epub 2022 Feb 3. Annu Rev Immunol. 2022. PMID: 35113730 Review.
Cited by
-
Phylogenetic and Evolutionary Analysis of Porcine Epidemic Diarrhea Virus in Guangxi Province, China, during 2020 and 2024.Viruses. 2024 Jul 14;16(7):1126. doi: 10.3390/v16071126. Viruses. 2024. PMID: 39066288 Free PMC article.
-
Genetic characteristics associated with the virulence of porcine epidemic diarrhea virus (PEDV) with a naturally occurring truncated ORF3 gene.Vet Res. 2024 Sep 27;55(1):123. doi: 10.1186/s13567-024-01384-w. Vet Res. 2024. PMID: 39334484 Free PMC article.
-
A Plant-Derived Maternal Vaccine against Porcine Epidemic Diarrhea Protects Piglets through Maternally Derived Immunity.Vaccines (Basel). 2023 May 9;11(5):965. doi: 10.3390/vaccines11050965. Vaccines (Basel). 2023. PMID: 37243069 Free PMC article.
-
Human super antibody to viral RNA-dependent RNA polymerase produced by a modified Sortase self-cleave-bacteria surface display system.Microb Cell Fact. 2023 Dec 18;22(1):260. doi: 10.1186/s12934-023-02267-z. Microb Cell Fact. 2023. PMID: 38110987 Free PMC article.
-
Antibody-Dependent Enhancement: ″Evil″ Antibodies Favorable for Viral Infections.Viruses. 2022 Aug 8;14(8):1739. doi: 10.3390/v14081739. Viruses. 2022. PMID: 36016361 Free PMC article. Review.
References
-
- Stevenson G.W., Hoang H., Schwartz K.J., Burrough E.R., Sun D., Madson D., Cooper V.L., Pillatzki A., Gauger P., Schmitt B.J., et al. Emergence of porcine epidemic diarrhea virus in the United States: Clinical signs, lesions, and viral genomic sequences. J. Vet. Diagn. Invest. 2013;25:649–654. doi: 10.1177/1040638713501675. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

