In Silico Prediction and Validation of CB2 Allosteric Binding Sites to Aid the Design of Allosteric Modulators
- PMID: 35056767
- PMCID: PMC8781014
- DOI: 10.3390/molecules27020453
In Silico Prediction and Validation of CB2 Allosteric Binding Sites to Aid the Design of Allosteric Modulators
Abstract
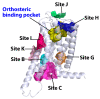
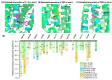
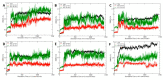
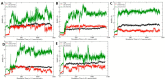
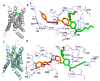
Although the 3D structures of active and inactive cannabinoid receptors type 2 (CB2) are available, neither the X-ray crystal nor the cryo-EM structure of CB2-orthosteric ligand-modulator has been resolved, prohibiting the drug discovery and development of CB2 allosteric modulators (AMs). In the present work, we mainly focused on investigating the potential allosteric binding site(s) of CB2. We applied different algorithms or tools to predict the potential allosteric binding sites of CB2 with the existing agonists. Seven potential allosteric sites can be observed for either CB2-CP55940 or CB2-WIN 55,212-2 complex, among which sites B, C, G and K are supported by the reported 3D structures of Class A GPCRs coupled with AMs. Applying our novel algorithm toolset-MCCS, we docked three known AMs of CB2 including Ec2la (C-2), trans-β-caryophyllene (TBC) and cannabidiol (CBD) to each site for further comparisons and quantified the potential binding residues in each allosteric binding site. Sequentially, we selected the most promising binding pose of C-2 in five allosteric sites to conduct the molecular dynamics (MD) simulations. Based on the results of docking studies and MD simulations, we suggest that site H is the most promising allosteric binding site. We plan to conduct bio-assay validations in the future.
Keywords: allosteric binding site; cannabinoid receptor 2; negative allosteric modulators; positive allosteric modulators.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Negative allosteric modulators of cannabinoid receptor 2: protein modeling, binding site identification and molecular dynamics simulations in the presence of an orthosteric agonist.J Biomol Struct Dyn. 2020 Jan;38(1):32-47. doi: 10.1080/07391102.2019.1567384. Epub 2019 Feb 5. J Biomol Struct Dyn. 2020. PMID: 30652534 Free PMC article.
-
Allosteric and orthosteric pharmacology of cannabidiol and cannabidiol-dimethylheptyl at the type 1 and type 2 cannabinoid receptors.Br J Pharmacol. 2019 May;176(10):1455-1469. doi: 10.1111/bph.14440. Epub 2018 Aug 10. Br J Pharmacol. 2019. PMID: 29981240 Free PMC article.
-
Modeling, molecular dynamics simulation, and mutation validation for structure of cannabinoid receptor 2 based on known crystal structures of GPCRs.J Chem Inf Model. 2014 Sep 22;54(9):2483-99. doi: 10.1021/ci5002718. Epub 2014 Sep 5. J Chem Inf Model. 2014. PMID: 25141027 Free PMC article.
-
Allosteric modulators targeting cannabinoid cb1 and cb2 receptors: implications for drug discovery.Future Med Chem. 2019 Aug;11(15):2019-2037. doi: 10.4155/fmc-2019-0005. Future Med Chem. 2019. PMID: 31517528 Review.
-
Leveraging allostery to improve G protein-coupled receptor (GPCR)-directed therapeutics: cannabinoid receptor 1 as discovery target.Expert Opin Drug Discov. 2016 Dec;11(12):1223-1237. doi: 10.1080/17460441.2016.1245289. Epub 2016 Oct 21. Expert Opin Drug Discov. 2016. PMID: 27712124 Free PMC article. Review.
Cited by
-
Allosteric regulation of kinase activity in living cells.Elife. 2023 Nov 9;12:RP90574. doi: 10.7554/eLife.90574. Elife. 2023. PMID: 37943025 Free PMC article.
-
Design, Synthesis, and Biological Activity of New CB2 Receptor Ligands: from Orthosteric and Allosteric Modulators to Dualsteric/Bitopic Ligands.J Med Chem. 2022 Jul 28;65(14):9918-9938. doi: 10.1021/acs.jmedchem.2c00582. Epub 2022 Jul 18. J Med Chem. 2022. PMID: 35849804 Free PMC article.
-
Cannabinoid type-2 receptors: An emerging target for regulating schizophrenia-relevant brain circuits.Front Neurosci. 2022 Aug 11;16:925792. doi: 10.3389/fnins.2022.925792. eCollection 2022. Front Neurosci. 2022. PMID: 36033626 Free PMC article. Review.
-
Targeting the endocannabinoid system: Structural determinants and molecular mechanism of allosteric modulation.Drug Discov Today. 2023 Jul;28(7):103615. doi: 10.1016/j.drudis.2023.103615. Epub 2023 May 11. Drug Discov Today. 2023. PMID: 37172889 Free PMC article. Review.
-
Integrative residue-intuitive machine learning and MD Approach to Unveil Allosteric Site and Mechanism for β2AR.Nat Commun. 2024 Sep 16;15(1):8130. doi: 10.1038/s41467-024-52399-y. Nat Commun. 2024. PMID: 39285201 Free PMC article.
References
-
- Feng Z., Alqarni M.H., Yang P., Tong Q., Chowdhury A., Wang L., Xie X.-Q. Modeling, Molecular Dynamics Simulation, and Mutation Validation for Structure of Cannabinoid Receptor 2 Based on Known Crystal Structures of GPCRs. J. Chem. Inf. Model. 2014;54:2483–2499. doi: 10.1021/ci5002718. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

