Complete Chloroplast Genome Sequence of Fagus longipetiolata Seemen (Fagaceae): Genome Structure, Adaptive Evolution, and Phylogenetic Relationships
- PMID: 35054485
- PMCID: PMC8778281
- DOI: 10.3390/life12010092
Complete Chloroplast Genome Sequence of Fagus longipetiolata Seemen (Fagaceae): Genome Structure, Adaptive Evolution, and Phylogenetic Relationships
Abstract
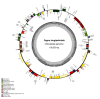
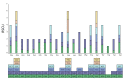
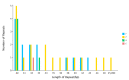
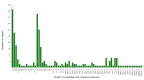
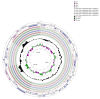
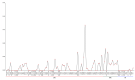
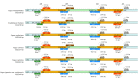
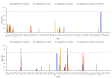
Fagus longipetiolata Seemen is a deciduous tree of the Fagus genus in Fagaceae, which is endemic to China. In this study, we successfully sequenced the cp genome of F. longipetiolata, compared the cp genomes of the Fagus genus, and reconstructed the phylogeny of Fagaceae. The results showed that the cp genome of F. longipetiolata was 158,350 bp, including a pair of inverted repeat (IRA and IRB) regions with a length of 25,894 bp each, a large single-copy (LSC) region of 87,671 bp, and a small single-copy (SSC) region of 18,891 bp. The genome encoded 131 unique genes, including 81 protein-coding genes, 37 transfer RNA genes (tRNAs), 8 ribosomal RNA genes (rRNAs), and 5 pseudogenes. In addition, 33 codons and 258 simple sequence repeats (SSRs) were identified. The cp genomes of Fagus were relatively conserved, especially the IR regions, which showed the best conservation, and no inversions or rearrangements were found. The five regions with the largest variations were the rps12, rpl32, ccsA, trnW-CCA, and rps3 genes, which spread over in LSC and SSC. The comparison of gene selection pressure indicated that purifying selection was the main selective pattern maintaining important biological functions in Fagus cp genomes. However, the ndhD, rpoA, and ndhF genes of F. longipetiolata were affected by positive selection. Phylogenetic analysis revealed that F. longipetiolata and F. engleriana formed a close relationship, which partially overlapped in their distribution in China. Our analysis of the cp genome of F. longipetiolata would provide important genetic information for further research into the classification, phylogeny and evolution of Fagus.
Keywords: F. longipetiolata; chloroplast genome; comparative analysis; phylogenetic analysis; purifying selection.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
The complete chloroplast genome of the medical plant Huperzia crispata from the Huperziaceae family: structure, comparative analysis, and phylogenetic relationships.Mol Biol Rep. 2022 Dec;49(12):11729-11741. doi: 10.1007/s11033-022-07979-w. Epub 2022 Oct 5. Mol Biol Rep. 2022. PMID: 36197623
-
The complete chloroplast genome of Fagus crenata (subgenus Fagus) and comparison with F. engleriana (subgenus Engleriana).PeerJ. 2019 Jun 7;7:e7026. doi: 10.7717/peerj.7026. eCollection 2019. PeerJ. 2019. PMID: 31211014 Free PMC article.
-
A systematic comparison of eight new plastome sequences from Ipomoea L.PeerJ. 2019 Mar 11;7:e6563. doi: 10.7717/peerj.6563. eCollection 2019. PeerJ. 2019. PMID: 30881765 Free PMC article.
-
Complete chloroplast genome sequence of Camellia sinensis: genome structure, adaptive evolution, and phylogenetic relationships.J Appl Genet. 2023 Sep;64(3):419-429. doi: 10.1007/s13353-023-00767-7. Epub 2023 Jun 28. J Appl Genet. 2023. PMID: 37380816
-
Comparative analysis of the complete chloroplast genomes of thirteen Bougainvillea cultivars from South China with implications for their genome structures and phylogenetic relationships.PLoS One. 2024 Sep 11;19(9):e0310091. doi: 10.1371/journal.pone.0310091. eCollection 2024. PLoS One. 2024. PMID: 39259741 Free PMC article.
Cited by
-
Phylogenomic analysis and dynamic evolution of chloroplast genomes of Clematis nannophylla.Sci Rep. 2024 Jul 2;14(1):15109. doi: 10.1038/s41598-024-65154-6. Sci Rep. 2024. PMID: 38956388 Free PMC article.
-
Comparison of chloroplast genomes and phylogenetic analysis of four species in Quercus section Cyclobalanopsis.Sci Rep. 2023 Oct 31;13(1):18731. doi: 10.1038/s41598-023-45421-8. Sci Rep. 2023. PMID: 37907468 Free PMC article.
-
Complete Plastome of Physalis angulata var. villosa, Gene Organization, Comparative Genomics and Phylogenetic Relationships among Solanaceae.Genes (Basel). 2022 Dec 5;13(12):2291. doi: 10.3390/genes13122291. Genes (Basel). 2022. PMID: 36553558 Free PMC article.
-
The complete chloroplast genome of Hibiscus syriacus using long-read sequencing: Comparative analysis to examine the evolution of the tribe Hibisceae.Front Plant Sci. 2023 Feb 2;14:1111968. doi: 10.3389/fpls.2023.1111968. eCollection 2023. Front Plant Sci. 2023. PMID: 36818825 Free PMC article.
-
Complete chloroplast genome of Lens lamottei reveals intraspecies variation among with Lens culinaris.Sci Rep. 2023 Sep 11;13(1):14959. doi: 10.1038/s41598-023-41287-y. Sci Rep. 2023. PMID: 37696838 Free PMC article.
References
-
- Guo K., Werger M.-J. Effect of prevailing monsoons on the distribution of beeches in continental East Asia. For. Ecol. Manag. 2010;259:2197–2203. doi: 10.1016/j.foreco.2009.11.034. - DOI
-
- Peters R. Ph.D. Thesis. Wageningen Agricultural University; Wageningen, The Netherlands: 1992. Ecology of Beech Forests in the Northern Hemisphere.
-
- Fang J., Lechowicz M.-J. Climatic limits for the present distribution of beech (Fagus L.) species in the world. J. Biogeogr. 2006;33:1804–1819. doi: 10.1111/j.1365-2699.2006.01533.x. - DOI
-
- Denk T. Phylogeny of Fagus L. (Fagaceae) based on morphological data. Plant Syst. Evol. 2003;240:55–81. doi: 10.1007/s00606-003-0018-x. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

