Correcting Differential Gene Expression Analysis for Cyto-Architectural Alterations in Substantia Nigra of Parkinson's Disease Patients Reveals Known and Potential Novel Disease-Associated Genes and Pathways
- PMID: 35053314
- PMCID: PMC8774027
- DOI: 10.3390/cells11020198
Correcting Differential Gene Expression Analysis for Cyto-Architectural Alterations in Substantia Nigra of Parkinson's Disease Patients Reveals Known and Potential Novel Disease-Associated Genes and Pathways
Abstract
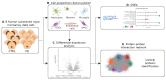
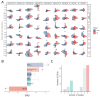
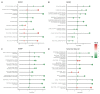
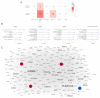
Several studies have analyzed gene expression profiles in the substantia nigra to better understand the pathological mechanisms causing Parkinson's disease (PD). However, the concordance between the identified gene signatures in these individual studies was generally low. This might have been caused by a change in cell type composition as loss of dopaminergic neurons in the substantia nigra pars compacta is a hallmark of PD. Through an extensive meta-analysis of nine previously published microarray studies, we demonstrated that a big proportion of the detected differentially expressed genes was indeed caused by cyto-architectural alterations due to the heterogeneity in the neurodegenerative stage and/or technical artefacts. After correcting for cell composition, we identified a common signature that deregulated the previously unreported ammonium transport, as well as known biological processes such as bioenergetic pathways, response to proteotoxic stress, and immune response. By integrating with protein interaction data, we shortlisted a set of key genes, such as LRRK2, PINK1, PRKN, and FBXO7, known to be related to PD, others with compelling evidence for their role in neurodegeneration, such as GSK3β, WWOX, and VPC, and novel potential players in the PD pathogenesis. Together, these data show the importance of accounting for cyto-architecture in these analyses and highlight the contribution of multiple cell types and novel processes to PD pathology, providing potential new targets for drug development.
Keywords: Parkinson’s disease; cyto-architecture; interactome analysis; meta-analysis; transcriptome analysis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Gene expression profiling of substantia nigra dopamine neurons: further insights into Parkinson's disease pathology.Brain. 2009 Jul;132(Pt 7):1795-809. doi: 10.1093/brain/awn323. Epub 2008 Dec 3. Brain. 2009. PMID: 19052140 Free PMC article.
-
An Integrated Network Analysis of mRNA and Gene Expression Profiles in Parkinson's Disease.Med Sci Monit. 2020 Mar 25;26:e920846. doi: 10.12659/MSM.920846. Med Sci Monit. 2020. PMID: 32210219 Free PMC article.
-
Construction of Parkinson's disease marker-based weighted protein-protein interaction network for prioritization of co-expressed genes.Gene. 2019 May 20;697:67-77. doi: 10.1016/j.gene.2019.02.026. Epub 2019 Feb 16. Gene. 2019. PMID: 30776463
-
Gene expression profiling of sporadic Parkinson's disease substantia nigra pars compacta reveals impairment of ubiquitin-proteasome subunits, SKP1A, aldehyde dehydrogenase, and chaperone HSC-70.Ann N Y Acad Sci. 2005 Aug;1053:356-75. doi: 10.1196/annals.1344.031. Ann N Y Acad Sci. 2005. PMID: 16179542 Review.
-
Convergence of signalling pathways in innate immune responses and genetic forms of Parkinson's disease.Neurobiol Dis. 2022 Jul;169:105721. doi: 10.1016/j.nbd.2022.105721. Epub 2022 Apr 8. Neurobiol Dis. 2022. PMID: 35405260 Review.
Cited by
-
A Systematic Review of Extracellular Matrix-Related Alterations in Parkinson's Disease.Brain Sci. 2024 May 21;14(6):522. doi: 10.3390/brainsci14060522. Brain Sci. 2024. PMID: 38928523 Free PMC article. Review.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

