53BP1-ACLY-SLBP-coordinated activation of replication-dependent histone biogenesis maintains genomic integrity
- PMID: 35037047
- PMCID: PMC8860602
- DOI: 10.1093/nar/gkab1300
53BP1-ACLY-SLBP-coordinated activation of replication-dependent histone biogenesis maintains genomic integrity
Abstract
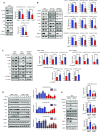
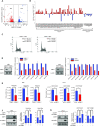
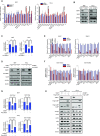
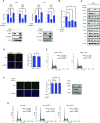
p53-binding protein 1 (53BP1) regulates the DNA double-strand break (DSB) repair pathway and maintains genomic integrity. Here we found that 53BP1 functions as a molecular scaffold for the nucleoside diphosphate kinase-mediated phosphorylation of ATP-citrate lyase (ACLY) which enhances the ACLY activity. This functional association is critical for promoting global histone acetylation and subsequent transcriptome-wide alterations in gene expression. Specifically, expression of a replication-dependent histone biogenesis factor, stem-loop binding protein (SLBP), is dependent upon 53BP1-ACLY-controlled acetylation at the SLBP promoter. This chain of regulation events carried out by 53BP1, ACLY, and SLBP is crucial for both quantitative and qualitative histone biogenesis as well as for the preservation of genomic integrity. Collectively, our findings reveal a previously unknown role for 53BP1 in coordinating replication-dependent histone biogenesis and highlight a DNA repair-independent function in the maintenance of genomic stability through a regulatory network that includes ACLY and SLBP.
© The Author(s) 2022. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


Similar articles
-
Nuclear Acetyl-CoA Production by ACLY Promotes Homologous Recombination.Mol Cell. 2017 Jul 20;67(2):252-265.e6. doi: 10.1016/j.molcel.2017.06.008. Epub 2017 Jul 6. Mol Cell. 2017. PMID: 28689661 Free PMC article.
-
Mammalian SIRT6 Represses Invasive Cancer Cell Phenotypes through ATP Citrate Lyase (ACLY)-Dependent Histone Acetylation.Genes (Basel). 2021 Sep 21;12(9):1460. doi: 10.3390/genes12091460. Genes (Basel). 2021. PMID: 34573442 Free PMC article.
-
Modulation of matrix metabolism by ATP-citrate lyase in articular chondrocytes.J Biol Chem. 2018 Aug 3;293(31):12259-12270. doi: 10.1074/jbc.RA118.002261. Epub 2018 Jun 21. J Biol Chem. 2018. PMID: 29929979 Free PMC article.
-
Regulation of DNA double-strand break repair pathway choice: a new focus on 53BP1.J Zhejiang Univ Sci B. 2021 Jan 15;22(1):38-46. doi: 10.1631/jzus.B2000306. J Zhejiang Univ Sci B. 2021. PMID: 33448186 Free PMC article. Review.
-
Roles for the DNA-PK complex and 53BP1 in protecting ends from resection during DNA double-strand break repair.J Radiat Res. 2020 Sep 8;61(5):718-726. doi: 10.1093/jrr/rraa053. J Radiat Res. 2020. PMID: 32779701 Free PMC article. Review.
Cited by
-
Histone locus bodies: a paradigm for how nuclear biomolecular condensates control cell cycle regulated gene expression.Nucleus. 2023 Dec;14(1):2293604. doi: 10.1080/19491034.2023.2293604. Epub 2023 Dec 14. Nucleus. 2023. PMID: 38095604 Free PMC article. Review.
References
-
- Ochs F., Somyajit K., Altmeyer M., Rask M.B., Lukas J., Lukas C.. 53BP1 fosters fidelity of homology-directed DNA repair. Nat. Struct. Mol. Biol. 2016; 23:714–721. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

