Host E3 ligase HUWE1 attenuates the proapoptotic activity of the MERS-CoV accessory protein ORF3 by promoting its ubiquitin-dependent degradation
- PMID: 35032548
- PMCID: PMC8755419
- DOI: 10.1016/j.jbc.2022.101584
Host E3 ligase HUWE1 attenuates the proapoptotic activity of the MERS-CoV accessory protein ORF3 by promoting its ubiquitin-dependent degradation
Abstract
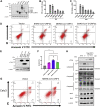
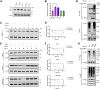
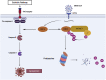
With the outbreak of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), coronaviruses have begun to attract great attention across the world. Of the known human coronaviruses, however, Middle East respiratory syndrome coronavirus (MERS-CoV) is the most lethal. Coronavirus proteins can be divided into three groups: nonstructural proteins, structural proteins, and accessory proteins. While the number of each of these proteins varies greatly among different coronaviruses, accessory proteins are most closely related to the pathogenicity of the virus. We found for the first time that the ORF3 accessory protein of MERS-CoV, which closely resembles the ORF3a proteins of severe acute respiratory syndrome coronavirus and SARS-CoV-2, has the ability to induce apoptosis in cells in a dose-dependent manner. Through bioinformatics analysis and validation, we revealed that ORF3 is an unstable protein and has a shorter half-life in cells compared to that of severe acute respiratory syndrome coronavirus and SARS-CoV-2 ORF3a proteins. After screening, we identified a host E3 ligase, HUWE1, that specifically induces MERS-CoV ORF3 protein ubiquitination and degradation through the ubiquitin-proteasome system. This results in the diminished ability of ORF3 to induce apoptosis, which might partially explain the lower spread of MERS-CoV compared to other coronaviruses. In summary, this study reveals a pathological function of MERS-CoV ORF3 protein and identifies a potential host antiviral protein, HUWE1, with an ability to antagonize MERS-CoV pathogenesis by inducing ORF3 degradation, thus enriching our knowledge of the pathogenesis of MERS-CoV and suggesting new targets and strategies for clinical development of drugs for MERS-CoV treatment.
Keywords: HUWE1; MERS-CoV; ORF3; apoptosis; degradation; ubiquitination.
Copyright © 2022 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures




Similar articles
-
UBR5 Acts as an Antiviral Host Factor against MERS-CoV via Promoting Ubiquitination and Degradation of ORF4b.J Virol. 2022 Sep 14;96(17):e0074122. doi: 10.1128/jvi.00741-22. Epub 2022 Aug 18. J Virol. 2022. PMID: 35980206 Free PMC article.
-
Antagonism of dsRNA-Induced Innate Immune Pathways by NS4a and NS4b Accessory Proteins during MERS Coronavirus Infection.mBio. 2019 Mar 26;10(2):e00319-19. doi: 10.1128/mBio.00319-19. mBio. 2019. PMID: 30914508 Free PMC article.
-
Middle East Respiratory Syndrome Coronavirus NS4b Protein Inhibits Host RNase L Activation.mBio. 2016 Mar 29;7(2):e00258. doi: 10.1128/mBio.00258-16. mBio. 2016. PMID: 27025250 Free PMC article.
-
Middle East respiratory syndrome coronavirus infection: virus-host cell interactions and implications on pathogenesis.Virol J. 2015 Dec 22;12:218. doi: 10.1186/s12985-015-0446-6. Virol J. 2015. PMID: 26690369 Free PMC article. Review.
-
A comparative review of pathogenesis and host innate immunity evasion strategies among the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), severe acute respiratory syndrome coronavirus (SARS-CoV) and the Middle East respiratory syndrome coronavirus (MERS-CoV).Arch Microbiol. 2021 Jul;203(5):1943-1951. doi: 10.1007/s00203-021-02265-y. Epub 2021 Mar 7. Arch Microbiol. 2021. PMID: 33682075 Free PMC article. Review.
Cited by
-
A Cullin 5-based complex serves as an essential modulator of ORF9b stability in SARS-CoV-2 replication.Signal Transduct Target Ther. 2024 Jun 28;9(1):159. doi: 10.1038/s41392-024-01874-5. Signal Transduct Target Ther. 2024. PMID: 38937432 Free PMC article.
-
Genetic diversity and molecular epidemiology of Middle East Respiratory Syndrome Coronavirus in dromedaries in Ethiopia, 2017-2020.Emerg Microbes Infect. 2023 Dec;12(1):e2164218. doi: 10.1080/22221751.2022.2164218. Emerg Microbes Infect. 2023. PMID: 36620913 Free PMC article.
-
A Perspective on Newly Emerging Proteolysis-Targeting Strategies in Antimicrobial Drug Discovery.Antibiotics (Basel). 2022 Nov 29;11(12):1717. doi: 10.3390/antibiotics11121717. Antibiotics (Basel). 2022. PMID: 36551374 Free PMC article. Review.
-
Coronavirus accessory protein ORF3 biology and its contribution to viral behavior and pathogenesis.iScience. 2023 Apr 21;26(4):106280. doi: 10.1016/j.isci.2023.106280. Epub 2023 Feb 28. iScience. 2023. PMID: 36945252 Free PMC article. Review.
-
UBR5 Acts as an Antiviral Host Factor against MERS-CoV via Promoting Ubiquitination and Degradation of ORF4b.J Virol. 2022 Sep 14;96(17):e0074122. doi: 10.1128/jvi.00741-22. Epub 2022 Aug 18. J Virol. 2022. PMID: 35980206 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

