Pseudomonas aeruginosa Biofilm Dispersion by the Human Atrial Natriuretic Peptide
- PMID: 35032112
- PMCID: PMC8895129
- DOI: 10.1002/advs.202103262
Pseudomonas aeruginosa Biofilm Dispersion by the Human Atrial Natriuretic Peptide
Abstract
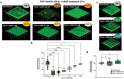
Pseudomonas aeruginosa biofilms cause chronic, antibiotic tolerant infections in wounds and lungs. Numerous recent studies demonstrate that bacteria can detect human communication compounds through specific sensor/receptor tools that modulate bacterial physiology. Consequently, interfering with these mechanisms offers an exciting opportunity to directly affect the infection process. It is shown that the human hormone Atrial Natriuretic Peptide (hANP) both prevents the formation of P. aeruginosa biofilms and strongly disperses established P. aeruginosa biofilms. This hANP action is dose-dependent with a strong effect at low nanomolar concentrations and takes effect in 30-120 min. Furthermore, although hANP has no antimicrobial effect, it acts as an antibiotic adjuvant. hANP enhances the antibiofilm action of antibiotics with diverse modes of action, allowing almost full biofilm eradication. The hANP effect requires the presence of the P. aeruginosa sensor AmiC and the AmiR antiterminator regulator, indicating a specific mode of action. These data establish the activation of the ami pathway as a potential mechanism for P. aeruginosa biofilm dispersion. hANP appears to be devoid of toxicity, does not enhance bacterial pathogenicity, and acts synergistically with antibiotics. These data show that hANP is a promising powerful antibiofilm weapon against established P. aeruginosa biofilms in chronic infections.
Keywords: Pseudomonas aeruginosa; ami pathway; antibiotics; bacterial adaptation; bacterial sensor; biofilm dispersion; hANP; natriuretic peptides.
© 2022 The Authors. Advanced Science published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
Pseudomonas aeruginosa Expresses a Functional Human Natriuretic Peptide Receptor Ortholog: Involvement in Biofilm Formation.mBio. 2015 Aug 25;6(4):e01033-15. doi: 10.1128/mBio.01033-15. mBio. 2015. PMID: 26307165 Free PMC article.
-
The natriuretic peptide receptor agonist osteocrin disperses Pseudomonas aeruginosa biofilm.Biofilm. 2023 May 19;5:100131. doi: 10.1016/j.bioflm.2023.100131. eCollection 2023 Dec. Biofilm. 2023. PMID: 37252226 Free PMC article.
-
Pseudomonas Quinolone Signal-Induced Outer Membrane Vesicles Enhance Biofilm Dispersion in Pseudomonas aeruginosa.mSphere. 2020 Nov 25;5(6):e01109-20. doi: 10.1128/mSphere.01109-20. mSphere. 2020. PMID: 33239369 Free PMC article.
-
Pathogenic factors of Pseudomonas aeruginosa - the role of biofilm in pathogenicity and as a target for phage therapy.Postepy Hig Med Dosw (Online). 2017 Feb 14;71(0):78-91. doi: 10.5604/01.3001.0010.3792. Postepy Hig Med Dosw (Online). 2017. PMID: 28258668 Review.
-
Bacteriophage therapy against Pseudomonas aeruginosa biofilms: a review.Ann Clin Microbiol Antimicrob. 2020 Sep 30;19(1):45. doi: 10.1186/s12941-020-00389-5. Ann Clin Microbiol Antimicrob. 2020. PMID: 32998720 Free PMC article. Review.
Cited by
-
Brain Natriuretic Peptide (BNP) Affects Growth and Stress Tolerance of Representatives of the Human Microbiome, Micrococcus luteus C01 and Alcaligenes faecalis DOS7.Biology (Basel). 2022 Jun 29;11(7):984. doi: 10.3390/biology11070984. Biology (Basel). 2022. PMID: 36101364 Free PMC article.
-
Carboxymethyl chitosan nanoparticle-modulated cationic hydrogels doped with copper ions for combating bacteria and facilitating wound healing.Front Bioeng Biotechnol. 2024 Sep 20;12:1429771. doi: 10.3389/fbioe.2024.1429771. eCollection 2024. Front Bioeng Biotechnol. 2024. PMID: 39372435 Free PMC article.
-
Biofilm formation: mechanistic insights and therapeutic targets.Mol Biomed. 2023 Dec 15;4(1):49. doi: 10.1186/s43556-023-00164-w. Mol Biomed. 2023. PMID: 38097907 Free PMC article. Review.
-
Combating Bacterial Biofilms: Current and Emerging Antibiofilm Strategies for Treating Persistent Infections.Antibiotics (Basel). 2023 Jun 3;12(6):1005. doi: 10.3390/antibiotics12061005. Antibiotics (Basel). 2023. PMID: 37370324 Free PMC article. Review.
-
Innovative Strategies to Overcome Antimicrobial Resistance and Tolerance.Microorganisms. 2022 Dec 21;11(1):16. doi: 10.3390/microorganisms11010016. Microorganisms. 2022. PMID: 36677308 Free PMC article. Review.
References
-
- Talwalkar J. S., Murray T. S., Clin. Chest Med. 2016, 37, 69. - PubMed
-
- Høiby N., Bjarnsholt T., Moser C., Bassi G. L., Coenye T., Donelli G., Hall‐Stoodley L., Holá V., Imbert C., Kirketerp‐Møller K., Lebeaux D., Oliver A., Ullmann A. J., Williams C., Clin. Microbiol. Infect. 2015, 21, S1. - PubMed
-
- Ciofu O., Rojo‐Molinero E., Macia M. D., Oliver A., APMIS 2017, 125, 304. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
