Human Cytomegalovirus Hijacks WD Repeat Domain 11 for Virion Assembly Compartment Formation and Virion Morphogenesis
- PMID: 35020472
- PMCID: PMC8906412
- DOI: 10.1128/JVI.01827-21
Human Cytomegalovirus Hijacks WD Repeat Domain 11 for Virion Assembly Compartment Formation and Virion Morphogenesis
Abstract
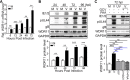
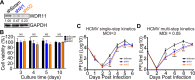
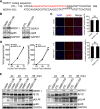
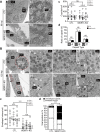
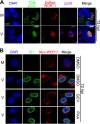
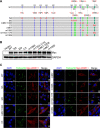
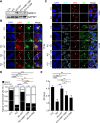
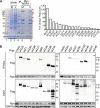
Human cytomegalovirus (HCMV) has a large (∼235 kb) genome with more than 200 predicted open reading frames that exploits numerous cellular factors to facilitate its replication. A key feature of HCMV-infected cells is the emergence of a distinctive membranous cytoplasmic compartment termed the virion assembly compartment (vAC). Here, we report that host protein WD repeat domain 11 (WDR11) plays a key role in vAC formation and virion morphogenesis. We found that WDR11 was upregulated at both mRNA and protein levels during HCMV infection. At the late stage of HCMV replication, WDR11 relocated to the vAC and colocalized with markers of the trans-Golgi network (TGN) and vAC. Depletion of WDR11 hindered HCMV-induced membrane reorganization of the Golgi and TGN, altered vAC formation, and impaired HCMV secondary envelopment and virion morphogenesis. Further, motifs critical for the localization of WDR11 in TGN were identified by alanine-scanning mutagenesis. Mutation of these motifs led to WDR11 mislocation outside the TGN and loss of vAC formation. Taken together, these data indicate that host protein WDR11 is required for efficient viral replication at the stage of virion assembly, possibly by facilitating the remodeling of the endomembrane system for vAC formation and virion morphogenesis. IMPORTANCE During the late phase of human cytomegalovirus (HCMV) infection, the endomembrane system is dramatically reorganized, resulting in the formation of a unique structure termed the virion assembly compartment (vAC), which is critical for the assembly of infectious virions. The mechanism of HCMV-induced vAC formation is still not fully understood. In this report, we identified a host factor, WDR11, that plays an important role in vAC formation. Our findings argue that WDR11 contributes to the relocation of the Golgi and trans-Golgi network to the vAC, a membrane reorganization process that appears to be required for efficient virion maturation. The present work provides new insights into the vAC formation and HCMV virion morphogenesis and a potential novel target for antiviral treatment.
Keywords: WDR11; human cytomegalovirus; virion assembly compartment; virion morphogenesis; virion structure.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Localization of the WD repeat-containing protein 5 to the Virion Assembly Compartment Facilitates Human Cytomegalovirus Assembly.J Virol. 2021 Mar 25;95(8):e02101-20. doi: 10.1128/JVI.02101-20. Epub 2021 Jan 27. J Virol. 2021. PMID: 33504601 Free PMC article.
-
Human cytomegalovirus exploits interferon-induced transmembrane proteins to facilitate morphogenesis of the virion assembly compartment.J Virol. 2015 Mar;89(6):3049-61. doi: 10.1128/JVI.03416-14. Epub 2014 Dec 31. J Virol. 2015. PMID: 25552713 Free PMC article.
-
Phosphorylation of Golgi Peripheral Membrane Protein Grasp65 Is an Integral Step in the Formation of the Human Cytomegalovirus Cytoplasmic Assembly Compartment.mBio. 2016 Oct 4;7(5):e01554-16. doi: 10.1128/mBio.01554-16. mBio. 2016. PMID: 27703074 Free PMC article.
-
Host Cell Signatures of the Envelopment Site within Beta-Herpes Virions.Int J Mol Sci. 2022 Sep 1;23(17):9994. doi: 10.3390/ijms23179994. Int J Mol Sci. 2022. PMID: 36077391 Free PMC article. Review.
-
Tegument proteins of human cytomegalovirus.Microbiol Mol Biol Rev. 2008 Jun;72(2):249-65, table of contents. doi: 10.1128/MMBR.00040-07. Microbiol Mol Biol Rev. 2008. PMID: 18535146 Free PMC article. Review.
Cited by
-
The human cytomegalovirus decathlon: Ten critical replication events provide opportunities for restriction.Front Cell Dev Biol. 2022 Nov 25;10:1053139. doi: 10.3389/fcell.2022.1053139. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36506089 Free PMC article. Review.
-
Coordination of canonical and noncanonical Hedgehog signalling pathways mediated by WDR11 during primordial germ cell development.Sci Rep. 2023 Jul 29;13(1):12309. doi: 10.1038/s41598-023-38017-9. Sci Rep. 2023. PMID: 37516749 Free PMC article.
-
The WDR11 complex is a receptor for acidic-cluster-containing cargo proteins.Cell. 2024 Aug 8;187(16):4272-4288.e20. doi: 10.1016/j.cell.2024.06.024. Epub 2024 Jul 15. Cell. 2024. PMID: 39013469
References
-
- Cobbs CS, Harkins L, Samanta M, Gillespie GY, Bharara S, King PH, Nabors LB, Cobbs CG, Britt WJ. 2002. Human cytomegalovirus infection and expression in human malignant glioma. Cancer Res 62:3347–3350. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

