Identification of the Key Immune-Related Genes in Chronic Obstructive Pulmonary Disease Based on Immune Infiltration Analysis
- PMID: 35018096
- PMCID: PMC8742581
- DOI: 10.2147/COPD.S333251
Identification of the Key Immune-Related Genes in Chronic Obstructive Pulmonary Disease Based on Immune Infiltration Analysis
Abstract
Purpose: Chronic obstructive pulmonary disease (COPD) is a major cause of death and morbidity worldwide. A better understanding of new biomarkers for COPD patients and their complex mechanisms in the progression of COPD are needed.
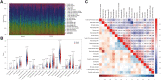
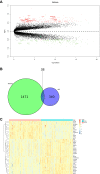
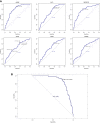
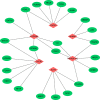
Methods: An algorithm was conducted to reveal the proportions of 22 subsets of immune cells in COPD samples. Differentially expressed immune-related genes (DE-IRGs) were obtained based on the differentially expressed genes (DEGs) of the GSE57148 dataset, and 1509 immune-related genes (IRGs) were downloaded from the ImmPort database. Functional enrichment analyses of DE-IRGs were conducted by Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses and Ingenuity Pathway Analysis (IPA). We defined the DE-IRGs that had correlations with immune cells as hub genes. The potential interactions among the hub genes were explored by a protein-protein interaction (PPI) network.
Results: The CIBERSORT results showed that lung tissue of COPD patients contained a greater number of resting NK cells, activated dendritic cells, and neutrophils than normal samples. However, the fractions of follicular helper T cells and resting dendritic cells were relatively lower. Thirty-eight DE-IRGs were obtained for further analysis. Functional enrichment analysis revealed that these DE-IRGs were significantly enriched in several immune-related biological processes and pathways. Notably, we also observed that DE-IRGs were associated with the coronavirus disease COVID-19 in the progression of COPD. After correlation analysis, six DE-IRGs associated with immune cells were considered hub genes, including AHNAK, SLIT2 TNFRRSF10C, CXCR1, CXCR2, and FCGR3B.
Conclusion: In the present study, we investigated immune-related genes as novel diagnostic biomarkers and explored the potential mechanism for COPD based on CIBERSORT analysis, providing a new understanding for COPD treatment.
Keywords: CIBERSORT; COPD; IRGs; chronic obstructive pulmonary disease; diagnosis; immune-related genes.
© 2022 Meng et al.
Conflict of interest statement
The authors report no conflicts of interest in this work.
Figures


Similar articles
-
Identification of Ferroptosis-Related Hub Genes and Their Association with Immune Infiltration in Chronic Obstructive Pulmonary Disease by Bioinformatics Analysis.Int J Chron Obstruct Pulmon Dis. 2022 May 24;17:1219-1236. doi: 10.2147/COPD.S348569. eCollection 2022. Int J Chron Obstruct Pulmon Dis. 2022. PMID: 35637927 Free PMC article.
-
Comprehensive transcriptomic analysis of immune-related genes in diabetic foot ulcers: New insights into mechanisms and therapeutic targets.Int Immunopharmacol. 2024 Sep 30;139:112638. doi: 10.1016/j.intimp.2024.112638. Epub 2024 Jul 28. Int Immunopharmacol. 2024. PMID: 39079197
-
Identification of novel biomarkers related to neutrophilic inflammation in COPD.Front Immunol. 2024 May 30;15:1410158. doi: 10.3389/fimmu.2024.1410158. eCollection 2024. Front Immunol. 2024. PMID: 38873611 Free PMC article.
-
Contribution of cuproptosis and Cu metabolism-associated genes to chronic obstructive pulmonary disease.J Cell Mol Med. 2023 Dec;27(24):4034-4044. doi: 10.1111/jcmm.17985. Epub 2023 Oct 6. J Cell Mol Med. 2023. PMID: 37801050 Free PMC article. Review.
-
Potential Mechanisms Between HF and COPD: New Insights From Bioinformatics.Curr Probl Cardiol. 2023 Mar;48(3):101539. doi: 10.1016/j.cpcardiol.2022.101539. Epub 2022 Dec 15. Curr Probl Cardiol. 2023. PMID: 36528207 Review.
Cited by
-
Does Chronic Obstructive Pulmonary Disease Originate from Different Cell Types?Am J Respir Cell Mol Biol. 2023 Nov;69(5):500-507. doi: 10.1165/rcmb.2023-0175PS. Am J Respir Cell Mol Biol. 2023. PMID: 37584669 Free PMC article.
-
Identification of COVID-19-Associated DNA Methylation Variations by Integrating Methylation Array and scRNA-Seq Data at Cell-Type Resolution.Genes (Basel). 2022 Jun 21;13(7):1109. doi: 10.3390/genes13071109. Genes (Basel). 2022. PMID: 35885892 Free PMC article.
-
Effectiveness of Immunotherapy in Non-Small Cell Lung Cancer Patients with a Diagnosis of COPD: Is This a Hidden Prognosticator for Survival and a Risk Factor for Immune-Related Adverse Events?Cancers (Basel). 2024 Mar 22;16(7):1251. doi: 10.3390/cancers16071251. Cancers (Basel). 2024. PMID: 38610929 Free PMC article. Review.
References
-
- Chen W, Hong Y-Q, Meng Z-L. Bioinformatics analysis of molecular mechanisms of chronic obstructive pulmonary disease. Eur Rev Med Pharmacol Sci. 2014;18(23):3557–3563. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

