Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies
- PMID: 35016194
- PMCID: PMC8866119
- DOI: 10.1038/s41586-021-04385-3
Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies
Abstract
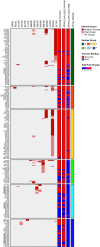
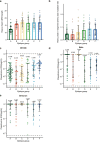
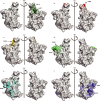
The SARS-CoV-2 B.1.1.529 (Omicron) variant contains 15 mutations of the receptor-binding domain (RBD). How Omicron evades RBD-targeted neutralizing antibodies requires immediate investigation. Here we use high-throughput yeast display screening1,2 to determine the profiles of RBD escaping mutations for 247 human anti-RBD neutralizing antibodies and show that the neutralizing antibodies can be classified by unsupervised clustering into six epitope groups (A-F)-a grouping that is highly concordant with knowledge-based structural classifications3-5. Various single mutations of Omicron can impair neutralizing antibodies of different epitope groups. Specifically, neutralizing antibodies in groups A-D, the epitopes of which overlap with the ACE2-binding motif, are largely escaped by K417N, G446S, E484A and Q493R. Antibodies in group E (for example, S309)6 and group F (for example, CR3022)7, which often exhibit broad sarbecovirus neutralizing activity, are less affected by Omicron, but a subset of neutralizing antibodies are still escaped by G339D, N440K and S371L. Furthermore, Omicron pseudovirus neutralization showed that neutralizing antibodies that sustained single mutations could also be escaped, owing to multiple synergetic mutations on their epitopes. In total, over 85% of the tested neutralizing antibodies were escaped by Omicron. With regard to neutralizing-antibody-based drugs, the neutralization potency of LY-CoV016, LY-CoV555, REGN10933, REGN10987, AZD1061, AZD8895 and BRII-196 was greatly undermined by Omicron, whereas VIR-7831 and DXP-604 still functioned at a reduced efficacy. Together, our data suggest that infection with Omicron would result in considerable humoral immune evasion, and that neutralizing antibodies targeting the sarbecovirus conserved region will remain most effective. Our results inform the development of antibody-based drugs and vaccines against Omicron and future variants.
© 2021. The Author(s).
Conflict of interest statement
X.S.X. and Y.C. are listed as inventors on a patent related to BD series antibodies and DXP-604 (PCT/CN2021/093305) under Peking University. X.S.X. and Y.C. are founders of Singlomics Biopharmaceuticals. The remaining authors declare no competing interests.
Figures




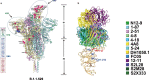






Comment in
-
Omicron, the great escape artist.Nat Rev Immunol. 2022 Feb;22(2):75. doi: 10.1038/s41577-022-00676-6. Nat Rev Immunol. 2022. PMID: 35017722 Free PMC article.
-
SARS-CoV-2 variant Omicron: currently the most complete "escapee" from neutralization by antibodies and vaccines.Signal Transduct Target Ther. 2022 Jan 28;7(1):28. doi: 10.1038/s41392-022-00880-9. Signal Transduct Target Ther. 2022. PMID: 35091532 Free PMC article. No abstract available.
Similar articles
-
Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift.Nature. 2022 Feb;602(7898):664-670. doi: 10.1038/s41586-021-04386-2. Epub 2021 Dec 23. Nature. 2022. PMID: 35016195 Free PMC article.
-
BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection.Nature. 2022 Aug;608(7923):593-602. doi: 10.1038/s41586-022-04980-y. Epub 2022 Jun 17. Nature. 2022. PMID: 35714668 Free PMC article.
-
Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2.Nature. 2022 Feb;602(7898):676-681. doi: 10.1038/s41586-021-04388-0. Epub 2021 Dec 23. Nature. 2022. PMID: 35016198
-
Mutations in the SARS-CoV-2 spike receptor binding domain and their delicate balance between ACE2 affinity and antibody evasion.Protein Cell. 2024 May 28;15(6):403-418. doi: 10.1093/procel/pwae007. Protein Cell. 2024. PMID: 38442025 Free PMC article. Review.
-
Analysis of the molecular mechanism of SARS-CoV-2 antibodies.Biochem Biophys Res Commun. 2021 Aug 20;566:45-52. doi: 10.1016/j.bbrc.2021.06.001. Epub 2021 Jun 5. Biochem Biophys Res Commun. 2021. PMID: 34116356 Free PMC article. Review.
Cited by
-
Act Early and at the Right Location: SARS-CoV-2 T Cell Kinetics and Tissue Localization.Int J Mol Sci. 2022 Sep 14;23(18):10679. doi: 10.3390/ijms231810679. Int J Mol Sci. 2022. PMID: 36142588 Free PMC article. Review.
-
Vaccine effectiveness against Delta, Omicron BA.1, and BA.2 in a highly vaccinated Asian setting: a test-negative design study.Clin Microbiol Infect. 2023 Jan;29(1):101-106. doi: 10.1016/j.cmi.2022.08.002. Epub 2022 Aug 24. Clin Microbiol Infect. 2023. PMID: 36028091 Free PMC article.
-
Clinical Evaluation of Direct Reverse Transcription PCR for Detection of SARS-CoV-2 Compared to Conventional RT-PCR in Patients with Positive Rapid Antigen Test Results during Circulation of Emerging Viral Variants.Diagnostics (Basel). 2023 Dec 14;13(24):3668. doi: 10.3390/diagnostics13243668. Diagnostics (Basel). 2023. PMID: 38132252 Free PMC article.
-
Distinct core glycan and O-glycoform utilization of SARS-CoV-2 Omicron variant Spike protein RBD revealed by top-down mass spectrometry.Chem Sci. 2022 Aug 31;13(36):10944-10949. doi: 10.1039/d2sc02132c. eCollection 2022 Sep 21. Chem Sci. 2022. PMID: 36320702 Free PMC article.
-
SARS-CoV-2 spike opening dynamics and energetics reveal the individual roles of glycans and their collective impact.Commun Biol. 2022 Nov 3;5(1):1170. doi: 10.1038/s42003-022-04138-6. Commun Biol. 2022. PMID: 36329138 Free PMC article.
References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

