[Identification of potential hub genes of Alzheimer's disease by weighted gene co-expression network analysis]
- PMID: 35012905
- PMCID: PMC8752417
- DOI: 10.12122/j.issn.1673-4254.2021.12.01
[Identification of potential hub genes of Alzheimer's disease by weighted gene co-expression network analysis]
Abstract
Objective: To investigate the differential expression gene modules and hub genes associated with Alzheimer's disease (AD) by weighted gene co-expression network analysis (WGCNA) and annotate the biological functions of these modules.
Methods: We downloaded transcriptome sequencing data from the GEO database, and according to the correlation of the genes, a gene co-expression network was constructed with the parameter setting of β=8 and a correlation coefficient threshold of 0.85. Pearson correlation test was used to calculate the correlation between the module genes and clinical traits to screen the gene modules significantly associated with AD and identify the hub genes according to the connectivity within modules. GO functional enrichment analysis and KEGG pathway analysis were used to annotate the functions of the modules. A cell model of AD was established in SH-SY5Y cells by Aβ1-42 treatment, and the mRNA expression levels of the hub genes were compared between the Aβ1-42-treated cells and the control cells.
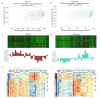
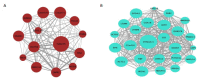
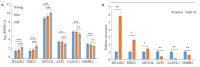
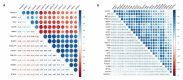
Results: Ten gene co-expression modules were constructed based on the correlations of gene expression, in which the brown (r=0.66, P < 0.001) and turquoise modules (r=-0.68, P < 0.001) were significantly correlated with the AD group. Forty-eight genes were identified as the hub genes in the co-expression network. Function annotation revealed that the genes in both modules were mainly enriched in DNA damage and repair pathways and metabolism-related pathways. Differential expression analysis of the genes revealed that the genes DNASE1, TEKT2 and MTSS1L were highly expressed while ACP2, LANCL2 and GMPR2 were lowly expressed in AD group. The results of cell experiment confirmed the up-regulation of DNASE1, TEKT2 and MTSS1L genes and the down-regulation of ACP2, LANCL2, and GMPR2 in Aβ1-42-treated SH-SY5Y cells (P < 0.01).
Conclusion: The brown and turquoise modules are closely correlated with AD. The hub genes including MTSS1L, GMPR2, ACP2, ACTG1 and LANCL2 selected from the modules may participate in AD pathogenesis by regulating DNA damage and repair.
目的: 采用加权基因共表达网络分析(WGCNA)探索阿尔茨海默病(AD)相关的差异基因模块及其枢纽基因,并对差异基因模块进行生物功能注释。
方法: 从GEO数据库下载转录组测序数据,根据基因的相关性,当关联系数阈值设定为0.85时,参数β=8,以此构建基因共表达网络;采用Pearson相关性检验计算模块基因与临床表型相关性,筛选出与AD显著相关的基因模块,根据模块内的连接性筛选枢纽基因;利用GO功能富集分析和KEGG通路分析对模块进行功能注释。进一步建立β-淀粉样蛋白(Aβ1-42)诱导SH-SY5Y细胞损伤模型,在模型组和对照组中检测枢纽基因的表达水平。结果根据基因表达的相关性,共构建了10个基因共表达模块,其中brown和turquoise模块与AD组显著相关(brown:r=0.66,p < 0.001;turquoise:r=-0.68,P < 0.001);
结果: 显示48个基因在共表达网络中处于核心地位;通过生物注释功能发现,两模块中的基因主要富集在DNA损伤修复通路和代谢相关通路等生物学过程中。基因的差异表达分析显示,DNASE1、TEKT2、MTSS1L等基因在AD组中高表达,ACP2、LANCL2、GMPR2等基因在AD组中低表达;体外实验进一步验证了在Aβ1-42诱导的SH-SY5Y细胞损伤过程中DNASE1、TEKT2、MTSS1L表达上调(P < 0.01),ACP2、LANCL2、GMPR2表达下调(P < 0.01)。
结论: brown和turquoise模块与AD高度相关,并从模块中筛选出MTSS1L、GMPR2、ACP2、ACTG1、LANCL2等枢纽基因,可能通过调节DNA损伤和修复参与AD发病机制。
Keywords: Alzheimer's disease; DNA damage and repair; hub genes; weighted gene co-expression network analysis.
Figures





Similar articles
-
Using weighted gene co-expression network analysis (WGCNA) to identify the hub genes related to hypoxic adaptation in yak (Bos grunniens).Genes Genomics. 2021 Oct;43(10):1231-1246. doi: 10.1007/s13258-021-01137-5. Epub 2021 Aug 2. Genes Genomics. 2021. PMID: 34338989
-
Identification of Potential Therapeutic Targets of Alzheimer's Disease By Weighted Gene Co-Expression Network Analysis.Chin Med Sci J. 2020 Dec 31;35(4):330-341. doi: 10.24920/003695. Chin Med Sci J. 2020. PMID: 33413749
-
Weighted gene co-expression network analysis revealed key biomarkers associated with the diagnosis of hypertrophic cardiomyopathy.Hereditas. 2020 Oct 24;157(1):42. doi: 10.1186/s41065-020-00155-9. Hereditas. 2020. PMID: 33099311 Free PMC article.
-
Identification of crosstalk genes and immune characteristics between Alzheimer's disease and atherosclerosis.Front Immunol. 2024 Aug 12;15:1443464. doi: 10.3389/fimmu.2024.1443464. eCollection 2024. Front Immunol. 2024. PMID: 39188714 Free PMC article.
-
Application of Weighted Gene Co-Expression Network Analysis to Explore the Key Genes in Alzheimer's Disease.J Alzheimers Dis. 2018;65(4):1353-1364. doi: 10.3233/JAD-180400. J Alzheimers Dis. 2018. PMID: 30124448 Free PMC article.
Cited by
-
Comparison of the Amyloid Plaque Proteome in Down Syndrome, Early-Onset Alzheimer's Disease and Late-Onset Alzheimer's Disease.Res Sq [Preprint]. 2024 Jul 15:rs.3.rs-4469045. doi: 10.21203/rs.3.rs-4469045/v1. Res Sq. 2024. Update in: Acta Neuropathol. 2025 Jan 18;149(1):9. doi: 10.1007/s00401-025-02844-z PMID: 39070643 Free PMC article. Updated. Preprint.
References
-
- Serrano-Pozo A, Frosch MP, Masliah E, et al. Neuropathological alterations in Alzheimer disease. http://www.onacademic.com/detail/journal_1000036512289110_2160.html. Cold Spring Harb Perspect Med. 2011;1(1):a006189–97. [Serrano-Pozo A, Frosch MP, Masliah E, et al. Neuropathological alterations in Alzheimer disease[J]. Cold Spring Harb Perspect Med, 2011, 1(1): a006189-97.] - PMC - PubMed
-
- Arnone MI, Davidson EH. The hardwiring of development: organization and function of genomic regulatory systems. Development. 1997;124(10):1851–64. doi: 10.1242/dev.124.10.1851. [Arnone MI, Davidson EH. The hardwiring of development: organization and function of genomic regulatory systems[J]. Development, 1997, 124(10): 1851-64.] - DOI - PubMed
-
- Miklos GL, Rubin GM. The role of the genome project in determining gene function: insights from model organisms. Cell. 1996;86(4):521–9. doi: 10.1016/S0092-8674(00)80126-9. [Miklos GL, Rubin GM. The role of the genome project in determining gene function: insights from model organisms[J]. Cell, 1996, 86(4): 521-9.] - DOI - PubMed
-
- Chen Y, Zhu J, Lum PY, et al. Variations in DNA elucidate molecular networks that cause disease. Nature. 2008;452(7186):429–35. doi: 10.1038/nature06757. [Chen Y, Zhu J, Lum PY, et al. Variations in DNA elucidate molecular networks that cause disease[J]. Nature, 2008, 452(7186): 429-35.] - DOI - PMC - PubMed
-
- Schadt EE, Lamb J, Yang X, et al. An integrative genomics approach to infer causal associations between gene expression and disease. Nat Genet. 2005;37(7):710–7. doi: 10.1038/ng1589. [Schadt EE, Lamb J, Yang X, et al. An integrative genomics approach to infer causal associations between gene expression and disease[J]. Nat Genet, 2005, 37(7): 710-7.] - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
