The importance of enhancer methylation for epigenetic regulation of tumorigenesis in squamous lung cancer
- PMID: 34987166
- PMCID: PMC8813945
- DOI: 10.1038/s12276-021-00718-4
The importance of enhancer methylation for epigenetic regulation of tumorigenesis in squamous lung cancer
Abstract
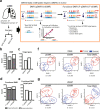
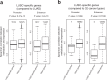
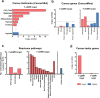
Lung squamous cell carcinoma (LUSC) is a subtype of non-small cell lung cancer (NSCLC). LUSC occurs at the bronchi, shows a squamous appearance, and often occurs in smokers. To determine the epigenetic regulatory mechanisms of tumorigenesis, we performed a genome-wide analysis of DNA methylation in tumor and adjacent normal tissues from LUSC patients. With the Infinium Methylation EPIC Array, > 850,000 CpG sites, including ~350,000 CpG sites for enhancer regions, were profiled, and the differentially methylated regions (DMRs) overlapping promoters (pDMRs) and enhancers (eDMRs) between tumor and normal tissues were identified. Dimension reduction based on DMR profiles revealed that eDMRs alone and not pDMRs alone can differentiate tumors from normal tissues with the equivalent performance of total DMRs. We observed a stronger negative correlation of LUSC-specific gene expression with methylation for enhancers than promoters. Target genes of eDMRs rather than pDMRs were found to be enriched for tumor-associated genes and pathways. Furthermore, DMR methylation associated with immune infiltration was more frequently observed among enhancers than promoters. Our results suggest that methylation of enhancer regions rather than promoters play more important roles in epigenetic regulation of tumorigenesis and immune infiltration in LUSC.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Genome-wide identification of differentially methylated promoters and enhancers associated with response to anti-PD-1 therapy in non-small cell lung cancer.Exp Mol Med. 2020 Sep;52(9):1550-1563. doi: 10.1038/s12276-020-00493-8. Epub 2020 Sep 2. Exp Mol Med. 2020. PMID: 32879421 Free PMC article.
-
Specific Lung Squamous Cell Carcinoma Prognosis-Subtype Distinctions Based on DNA Methylation Patterns.Med Sci Monit. 2021 Mar 4;27:e929524. doi: 10.12659/MSM.929524. Med Sci Monit. 2021. PMID: 33661858 Free PMC article.
-
Blood FOLR3 methylation dysregulations and heterogeneity in non-small lung cancer highlight its strong associations with lung squamous carcinoma.Respir Res. 2024 Jan 25;25(1):59. doi: 10.1186/s12931-024-02691-8. Respir Res. 2024. PMID: 38273401 Free PMC article.
-
Integrative analysis of DNA methylation-driven genes for the prognosis of lung squamous cell carcinoma using MethylMix.Int J Med Sci. 2020 Mar 5;17(6):773-786. doi: 10.7150/ijms.43272. eCollection 2020. Int J Med Sci. 2020. PMID: 32218699 Free PMC article.
-
Identification of Key DNA methylation sites related to differentially expressed genes in Lung squamous cell carcinoma.Comput Biol Med. 2023 Dec;167:107615. doi: 10.1016/j.compbiomed.2023.107615. Epub 2023 Oct 25. Comput Biol Med. 2023. PMID: 37918267
Cited by
-
Plasma Cell-Free Tumor Methylome as a Biomarker in Solid Tumors: Biology and Applications.Curr Oncol. 2024 Jan 13;31(1):482-500. doi: 10.3390/curroncol31010033. Curr Oncol. 2024. PMID: 38248118 Free PMC article. Review.
-
EpiMix is an integrative tool for epigenomic subtyping using DNA methylation.Cell Rep Methods. 2023 Jun 22;3(7):100515. doi: 10.1016/j.crmeth.2023.100515. eCollection 2023 Jul 24. Cell Rep Methods. 2023. PMID: 37533639 Free PMC article.
-
Whole genome methylation sequencing in blood identifies extensive differential DNA methylation in late-onset dementia due to Alzheimer's disease.Alzheimers Dement. 2024 Feb;20(2):1050-1062. doi: 10.1002/alz.13514. Epub 2023 Oct 19. Alzheimers Dement. 2024. PMID: 37856321 Free PMC article.
-
Cis- and trans-eQTM analysis reveals novel epigenetic and transcriptomic immune markers of atopic asthma in airway epithelium.J Allergy Clin Immunol. 2023 Oct;152(4):887-898. doi: 10.1016/j.jaci.2023.05.018. Epub 2023 Jun 2. J Allergy Clin Immunol. 2023. PMID: 37271320 Free PMC article.
-
Reshaping the tumour immune microenvironment in solid tumours via tumour cell and immune cell DNA methylation: from mechanisms to therapeutics.Br J Cancer. 2023 Jul;129(1):24-37. doi: 10.1038/s41416-023-02292-0. Epub 2023 Apr 28. Br J Cancer. 2023. PMID: 37117649 Free PMC article. Review.
References
-
- Travis WD, et al. The 2015 World Health Organization classification of lung tumors: impact of genetic, clinical and radiologic advances since the 2004 classification. J. Thorac. Oncol. 2015;10:1243–1260. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

