DNMT1 regulates the timing of DNA methylation by DNMT3 in an enzymatic activity-dependent manner in mouse embryonic stem cells
- PMID: 34986190
- PMCID: PMC8730390
- DOI: 10.1371/journal.pone.0262277
DNMT1 regulates the timing of DNA methylation by DNMT3 in an enzymatic activity-dependent manner in mouse embryonic stem cells
Abstract
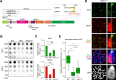
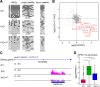
DNA methylation (DNAme; 5-methylcytosine, 5mC) plays an essential role in mammalian development, and the 5mC profile is regulated by a balance of opposing enzymatic activities: DNA methyltransferases (DNMTs) and Ten-eleven translocation dioxygenases (TETs). In mouse embryonic stem cells (ESCs), de novo DNAme by DNMT3 family enzymes, demethylation by the TET-mediated conversion of 5mC to 5-hydroxymethylation (5hmC), and maintenance of the remaining DNAme by DNMT1 are actively repeated throughout cell cycles, dynamically forming a constant 5mC profile. Nevertheless, the detailed mechanism and physiological significance of this active cyclic DNA modification in mouse ESCs remain unclear. Here by visualizing the localization of DNA modifications on metaphase chromosomes and comparing whole-genome methylation profiles before and after the mid-S phase in ESCs lacking Dnmt1 (1KO ESCs), we demonstrated that in 1KO ESCs, DNMT3-mediated remethylation was interrupted during and after DNA replication. This results in a marked asymmetry in the distribution of 5hmC between sister chromatids at mitosis, with one chromatid being almost no 5hmC. When introduced in 1KO ESCs, the catalytically inactive form of DNMT1 (DNMT1CI) induced an increase in DNAme in pericentric heterochromatin and the DNAme-independent repression of IAPEz, a retrotransposon family, in 1KO ESCs. However, DNMT1CI could not restore the ability of DNMT3 to methylate unmodified dsDNA de novo in S phase in 1KO ESCs. Furthermore, during in vitro differentiation into epiblasts, 1KO ESCs expressing DNMT1CI showed an even stronger tendency to differentiate into the primitive endoderm than 1KO ESCs and were readily reprogrammed into the primitive streak via an epiblast-like cell state, reconfirming the importance of DNMT1 enzymatic activity at the onset of epiblast differentiation. These results indicate a novel function of DNMT1, in which DNMT1 actively regulates the timing and genomic targets of de novo methylation by DNMT3 in an enzymatic activity-dependent and independent manner, respectively.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
DNMT1 can induce primary germ layer differentiation through de novo DNA methylation.Genes Cells. 2024 Jul;29(7):549-566. doi: 10.1111/gtc.13130. Epub 2024 May 29. Genes Cells. 2024. PMID: 38811355 Free PMC article.
-
Chromosome-wide regulation of euchromatin-specific 5mC to 5hmC conversion in mouse ES cells and female human somatic cells.Chromosome Res. 2012 Oct;20(7):837-48. doi: 10.1007/s10577-012-9317-9. Epub 2012 Oct 31. Chromosome Res. 2012. PMID: 23111490 Free PMC article.
-
Widespread recovery of methylation at gametic imprints in hypomethylated mouse stem cells following rescue with DNMT3A2.Epigenetics Chromatin. 2016 Nov 22;9:53. doi: 10.1186/s13072-016-0104-2. eCollection 2016. Epigenetics Chromatin. 2016. PMID: 27895716 Free PMC article.
-
TET methylcytosine oxidases: new insights from a decade of research.J Biosci. 2020;45:21. J Biosci. 2020. PMID: 31965999 Free PMC article. Review.
-
Local chromatin microenvironment determines DNMT activity: from DNA methyltransferase to DNA demethylase or DNA dehydroxymethylase.Epigenetics. 2015;10(8):671-6. doi: 10.1080/15592294.2015.1062204. Epigenetics. 2015. PMID: 26098813 Free PMC article. Review.
Cited by
-
5-methylcytosine turnover: Mechanisms and therapeutic implications in cancer.Front Mol Biosci. 2022 Aug 17;9:976862. doi: 10.3389/fmolb.2022.976862. eCollection 2022. Front Mol Biosci. 2022. PMID: 36060265 Free PMC article. Review.
-
DNMT1 can induce primary germ layer differentiation through de novo DNA methylation.Genes Cells. 2024 Jul;29(7):549-566. doi: 10.1111/gtc.13130. Epub 2024 May 29. Genes Cells. 2024. PMID: 38811355 Free PMC article.
-
Milk Exosomal microRNAs: Postnatal Promoters of β Cell Proliferation but Potential Inducers of β Cell De-Differentiation in Adult Life.Int J Mol Sci. 2022 Sep 29;23(19):11503. doi: 10.3390/ijms231911503. Int J Mol Sci. 2022. PMID: 36232796 Free PMC article. Review.
-
The molecular basis of cell memory in mammals: The epigenetic cycle.Sci Adv. 2024 Mar;10(9):eadl3188. doi: 10.1126/sciadv.adl3188. Epub 2024 Feb 28. Sci Adv. 2024. PMID: 38416817 Free PMC article. Review.
References
-
- Berkyurek AC, Suetake I, Arita K, Takeshita K, Nakagawa A, Shirakawa M, et al.. The DNA methyltransferase Dnmt1 directly interacts with the SET and RING finger-associated (SRA) domain of the multifunctional protein Uhrf1 to facilitate accession of the catalytic center to hemi-methylated DNA. J Biol Chem. 2014;289: 379–386. doi: 10.1074/jbc.M113.523209 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

