Type I and II PRMTs inversely regulate post-transcriptional intron detention through Sm and CHTOP methylation
- PMID: 34984976
- PMCID: PMC8765754
- DOI: 10.7554/eLife.72867
Type I and II PRMTs inversely regulate post-transcriptional intron detention through Sm and CHTOP methylation
Abstract
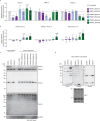
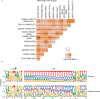
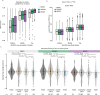
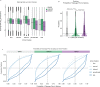
Protein arginine methyltransferases (PRMTs) are required for the regulation of RNA processing factors. Type I PRMT enzymes catalyze mono- and asymmetric dimethylation; Type II enzymes catalyze mono- and symmetric dimethylation. To understand the specific mechanisms of PRMT activity in splicing regulation, we inhibited Type I and II PRMTs and probed their transcriptomic consequences. Using the newly developed Splicing Kinetics and Transcript Elongation Rates by Sequencing (SKaTER-seq) method, analysis of co-transcriptional splicing demonstrated that PRMT inhibition resulted in altered splicing rates. Surprisingly, co-transcriptional splicing kinetics did not correlate with final changes in splicing of polyadenylated RNA. This was particularly true for retained introns (RI). By using actinomycin D to inhibit ongoing transcription, we determined that PRMTs post-transcriptionally regulate RI. Subsequent proteomic analysis of both PRMT-inhibited chromatin and chromatin-associated polyadenylated RNA identified altered binding of many proteins, including the Type I substrate, CHTOP, and the Type II substrate, SmB. Targeted mutagenesis of all methylarginine sites in SmD3, SmB, and SmD1 recapitulated splicing changes seen with Type II PRMT inhibition, without disrupting snRNP assembly. Similarly, mutagenesis of all methylarginine sites in CHTOP recapitulated the splicing changes seen with Type I PRMT inhibition. Examination of subcellular fractions further revealed that RI were enriched in the nucleoplasm and chromatin. Taken together, these data demonstrate that, through Sm and CHTOP arginine methylation, PRMTs regulate the post-transcriptional processing of nuclear, detained introns.
Keywords: A549; PRMT1; PRMT5; RNA processing; adma; cancer biology; carm1; cell biology; detained introns; di; mass spectrometry; mma; post-transcriptional processing; post-translational modifications; prmt1; prmt4; prmt5; prmts; protein arginine methyltransferases; ptms; retained detained introns; rme1; rme2a; rme2s; sdma; snRNP; splicing; transcription.
© 2022, Maron et al.
Conflict of interest statement
MM, AC, VG, JR, SS, CQ, MG, DS No competing interests declared
Figures






Similar articles
-
Productive mRNA Chromatin Escape is Promoted by PRMT5 Methylation of SNRPB.bioRxiv [Preprint]. 2024 Aug 12:2024.08.09.607355. doi: 10.1101/2024.08.09.607355. bioRxiv. 2024. PMID: 39149374 Free PMC article. Preprint.
-
Independent transcriptomic and proteomic regulation by type I and II protein arginine methyltransferases.iScience. 2021 Aug 11;24(9):102971. doi: 10.1016/j.isci.2021.102971. eCollection 2021 Sep 24. iScience. 2021. PMID: 34505004 Free PMC article.
-
Modulating the expression of Chtop, a versatile regulator of gene-specific transcription and mRNA export.RNA Biol. 2018;15(7):849-855. doi: 10.1080/15476286.2018.1465795. Epub 2018 May 11. RNA Biol. 2018. PMID: 29683372 Free PMC article. Review.
-
Simultaneous ablation of prmt-1 and prmt-5 abolishes asymmetric and symmetric arginine dimethylations in Caenorhabditis elegans.J Biochem. 2017 Jun 1;161(6):521-527. doi: 10.1093/jb/mvw101. J Biochem. 2017. PMID: 28158808
-
Cellular consequences of arginine methylation.Cell Mol Life Sci. 2019 Aug;76(15):2933-2956. doi: 10.1007/s00018-019-03140-2. Epub 2019 May 17. Cell Mol Life Sci. 2019. PMID: 31101937 Free PMC article. Review.
Cited by
-
Tudor-dimethylarginine interactions: the condensed version.Trends Biochem Sci. 2023 Aug;48(8):689-698. doi: 10.1016/j.tibs.2023.04.003. Epub 2023 May 6. Trends Biochem Sci. 2023. PMID: 37156649 Free PMC article. Review.
-
PRMT2 silencing regulates macrophage polarization through activation of STAT1 or inhibition of STAT6.BMC Immunol. 2024 Jan 3;25(1):1. doi: 10.1186/s12865-023-00593-w. BMC Immunol. 2024. PMID: 38172698 Free PMC article.
-
Effectors and effects of arginine methylation.Biochem Soc Trans. 2023 Apr 26;51(2):725-734. doi: 10.1042/BST20221147. Biochem Soc Trans. 2023. PMID: 37013969 Free PMC article.
-
Productive mRNA Chromatin Escape is Promoted by PRMT5 Methylation of SNRPB.bioRxiv [Preprint]. 2024 Aug 12:2024.08.09.607355. doi: 10.1101/2024.08.09.607355. bioRxiv. 2024. PMID: 39149374 Free PMC article. Preprint.
-
Competing Endogenous RNA (ceRNA) Networks and Splicing Switches in Cervical Cancer: HPV Oncogenesis, Clinical Significance and Therapeutic Opportunities.Microorganisms. 2022 Sep 16;10(9):1852. doi: 10.3390/microorganisms10091852. Microorganisms. 2022. PMID: 36144454 Free PMC article. Review.
References
-
- Bezzi M, Teo SX, Muller J, Mok WC, Sahu SK, Vardy LA, Bonday ZQ, Guccione E. Regulation of constitutive and alternative splicing by PRMT5 reveals a role for Mdm4 pre-mRNA in sensing defects in the spliceosomal machinery. Genes & Development. 2013;27:1903–1916. doi: 10.1101/gad.219899.113. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

