Protein synthesis, degradation, and energy metabolism in T cell immunity
- PMID: 34983947
- PMCID: PMC8891282
- DOI: 10.1038/s41423-021-00792-8
Protein synthesis, degradation, and energy metabolism in T cell immunity
Abstract
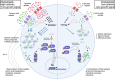
T cell activation, proliferation, and differentiation into effector and memory states involve massive remodeling of T cell size and molecular content and create a massive increase in demand for energy and amino acids. Protein synthesis is an energy- and resource-demanding process; as such, changes in T cell energy production are intrinsically linked to proteome remodeling. In this review, we discuss how protein synthesis and degradation change over the course of a T cell immune response and the crosstalk between these processes and T cell energy metabolism. We highlight how the use of high-resolution mass spectrometry to analyze T cell proteomes can improve our understanding of how these processes are regulated.
Keywords: Immunometabolism; Protein Translation; Protein degradation; Proteomics; T lymphocyte.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Emerging insights and challenges for understanding T cell function through the proteome.Front Immunol. 2022 Nov 16;13:1028366. doi: 10.3389/fimmu.2022.1028366. eCollection 2022. Front Immunol. 2022. PMID: 36466897 Free PMC article. Review.
-
Quantitative analysis of how Myc controls T cell proteomes and metabolic pathways during T cell activation.Elife. 2020 Feb 5;9:e53725. doi: 10.7554/eLife.53725. Elife. 2020. PMID: 32022686 Free PMC article.
-
Unraveling the Complex Interplay Between T Cell Metabolism and Function.Annu Rev Immunol. 2018 Apr 26;36:461-488. doi: 10.1146/annurev-immunol-042617-053019. Annu Rev Immunol. 2018. PMID: 29677474 Free PMC article. Review.
-
Mitochondrial Biogenesis and Proteome Remodeling Promote One-Carbon Metabolism for T Cell Activation.Cell Metab. 2016 Jul 12;24(1):104-17. doi: 10.1016/j.cmet.2016.06.007. Cell Metab. 2016. PMID: 27411012 Free PMC article.
-
The Ups and Downs of Metabolism during the Lifespan of a T Cell.Int J Mol Sci. 2020 Oct 27;21(21):7972. doi: 10.3390/ijms21217972. Int J Mol Sci. 2020. PMID: 33120978 Free PMC article. Review.
Cited by
-
Bisphenol A Disrupts Ribosome Function during Ovarian Development of Mice.Toxics. 2024 Aug 26;12(9):627. doi: 10.3390/toxics12090627. Toxics. 2024. PMID: 39330555 Free PMC article.
-
Proteostasis in T cell aging.Semin Immunol. 2023 Nov;70:101838. doi: 10.1016/j.smim.2023.101838. Epub 2023 Sep 12. Semin Immunol. 2023. PMID: 37708826 Free PMC article. Review.
-
Metabolic dysregulation impairs lymphocyte function during severe SARS-CoV-2 infection.Commun Biol. 2023 Apr 7;6(1):374. doi: 10.1038/s42003-023-04730-4. Commun Biol. 2023. PMID: 37029220 Free PMC article.
-
Optimization of ribosome utilization in Saccharomyces cerevisiae.PNAS Nexus. 2023 Mar 9;2(3):pgad074. doi: 10.1093/pnasnexus/pgad074. eCollection 2023 Mar. PNAS Nexus. 2023. PMID: 37007710 Free PMC article.
-
Proteomic and functional comparison between human induced and embryonic stem cells.Elife. 2024 Nov 14;13:RP92025. doi: 10.7554/eLife.92025. Elife. 2024. PMID: 39540879 Free PMC article.
References
-
- Marchingo JM, Kan A, Sutherland RM, Duffy KR, Wellard CJ, Belz GT, et al. T cell signaling. Antigen affinity, costimulation, and cytokine inputs sum linearly to amplify T cell expansion. Science. 2014;346:1123–7. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

