Computational Prediction of Biomarkers, Pathways, and New Target Drugs in the Pathogenesis of Immune-Based Diseases Regarding Kidney Transplantation Rejection
- PMID: 34975915
- PMCID: PMC8714745
- DOI: 10.3389/fimmu.2021.800968
Computational Prediction of Biomarkers, Pathways, and New Target Drugs in the Pathogenesis of Immune-Based Diseases Regarding Kidney Transplantation Rejection
Abstract
Background: The diagnosis of graft rejection in kidney transplantation (KT) patients is made by evaluating the histological characteristics of biopsy samples. The evolution of omics sciences and bioinformatics techniques has contributed to the advancement in searching and predicting biomarkers, pathways, and new target drugs that allow a more precise and less invasive diagnosis. The aim was to search for differentially expressed genes (DEGs) in patients with/without antibody-mediated rejection (AMR) and find essential cells involved in AMR, new target drugs, protein-protein interactions (PPI), and know their functional and biological analysis.
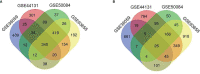
Material and methods: Four GEO databases of kidney biopsies of kidney transplantation with/without AMR were analyzed. The infiltrating leukocyte populations in the graft, new target drugs, protein-protein interactions (PPI), functional and biological analysis were studied by different bioinformatics tools.
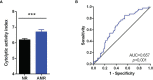
Results: Our results show DEGs and the infiltrating leukocyte populations in the graft. There is an increase in the expression of genes related to different stages of the activation of the immune system, antigenic presentation such as antibody-mediated cytotoxicity, or leukocyte migration during AMR. The importance of the IRF/STAT1 pathways of response to IFN in controlling the expression of genes related to humoral rejection. The genes of this biological pathway were postulated as potential therapeutic targets and biomarkers of AMR. These biological processes correlated showed the infiltration of NK cells and monocytes towards the allograft. Besides the increase in dendritic cell maturation, it plays a central role in mediating the damage suffered by the graft during AMR. Computational approaches to the search for new therapeutic uses of approved target drugs also showed that imatinib might theoretically be helpful in KT for the prevention and/or treatment of AMR.
Conclusion: Our results suggest the importance of the IRF/STAT1 pathways in humoral kidney rejection. NK cells and monocytes in graft damage have an essential role during rejection, and imatinib improves KT outcomes. Our results will have to be validated for the potential use of overexpressed genes as rejection biomarkers that can be used as diagnostic and prognostic markers and as therapeutic targets to avoid graft rejection in patients undergoing kidney transplantation.
Keywords: acute rejection; bioinformatics tool; biomarkers; kidney transplant; new target drugs.
Copyright © 2021 Alfaro, Martínez-Banaclocha, Llorente, Jimenez-Coll, Galián, Botella, Moya-Quiles, Parrado, Muro-Perez, Minguela, Legaz and Muro.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
CCR7 and CD48 as Predicted Targets in Acute Rejection Related to M1 Macrophage after Pediatric Kidney Transplantation.J Immunol Res. 2024 Jun 24;2024:6908968. doi: 10.1155/2024/6908968. eCollection 2024. J Immunol Res. 2024. PMID: 38957433 Free PMC article.
-
Bortezomib in late antibody-mediated kidney transplant rejection (BORTEJECT Study): study protocol for a randomized controlled trial.Trials. 2014 Apr 3;15:107. doi: 10.1186/1745-6215-15-107. Trials. 2014. PMID: 24708575 Free PMC article. Clinical Trial.
-
A Banff Component Scoring-based Histologic Assessment of Bortezomib-based Antibody-mediated Rejection Therapy.Transplantation. 2015 Aug;99(8):1691-9. doi: 10.1097/TP.0000000000000694. Transplantation. 2015. PMID: 25803498
-
Approaches for Controlling Antibody-Mediated Allograft Rejection Through Targeting B Cells.Front Immunol. 2021 Jul 1;12:682334. doi: 10.3389/fimmu.2021.682334. eCollection 2021. Front Immunol. 2021. PMID: 34276669 Free PMC article. Review.
-
Recent Advances on Biomarkers of Early and Late Kidney Graft Dysfunction.Int J Mol Sci. 2020 Jul 29;21(15):5404. doi: 10.3390/ijms21155404. Int J Mol Sci. 2020. PMID: 32751357 Free PMC article. Review.
Cited by
-
Dissecting the impact of molecular T-cell HLA mismatches in kidney transplant failure: A retrospective cohort study.Front Immunol. 2022 Nov 24;13:1067075. doi: 10.3389/fimmu.2022.1067075. eCollection 2022. Front Immunol. 2022. PMID: 36505483 Free PMC article.
-
Serum uric acid as a risk factor for rejection after deceased donor kidney transplantation: A mono-institutional analysis of paired kidneys.Front Immunol. 2022 Dec 12;13:973425. doi: 10.3389/fimmu.2022.973425. eCollection 2022. Front Immunol. 2022. PMID: 36578496 Free PMC article.
-
Monitoring of Serological, Cellular and Genomic Biomarkers in Transplantation, Computational Prediction Models and Role of Cell-Free DNA in Transplant Outcome.Int J Mol Sci. 2023 Feb 15;24(4):3908. doi: 10.3390/ijms24043908. Int J Mol Sci. 2023. PMID: 36835314 Free PMC article. Review.
-
MicroRNAs as Potential Graft Rejection or Tolerance Biomarkers and Their Dilemma in Clinical Routines Behaving like Devilish, Angelic, or Frightening Elements.Biomedicines. 2024 Jan 5;12(1):116. doi: 10.3390/biomedicines12010116. Biomedicines. 2024. PMID: 38255221 Free PMC article. Review.
-
Multi-omics Approach in Kidney Transplant: Lessons Learned from COVID-19 Pandemic.Curr Transplant Rep. 2023 Dec;10(4):173-187. doi: 10.1007/s40472-023-00410-8. Epub 2023 Aug 23. Curr Transplant Rep. 2023. PMID: 38152593 Free PMC article.
References
-
- Legaz I, Bernardo MV, Alfaro R, Martínez-Banaclocha H, Galián JA, Jimenez-Coll V, et al. . PCR Array Technology in Biopsy Samples Identifies Up-Regulated mTOR Pathway Genes as Potential Rejection Biomarkers After Kidney Transplantation. Front Med (2021) 0:547849. doi: 10.3389/FMED.2021.547849 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

